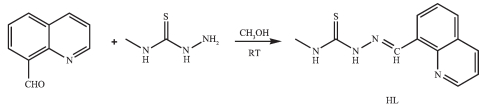
 Figure Scheme 1.
Synthetic route for the synthesis of HL
Figure Scheme 1.
Synthetic route for the synthesis of HL

喹啉-8-甲醛缩4-甲基氨基硫脲Ni(Ⅱ)/Zn(Ⅱ)/Cd(Ⅱ)/Cu(Ⅱ) 配合物的合成、结构和DNA结合性质
English
Syntheses, Crystal Structures and DNA-Binding Properties of Ni(Ⅱ)/Zn(Ⅱ)/Cd(Ⅱ)/Cu(Ⅱ) Complexes with 4-Methyl-1-((quinolin-8-yl) methylene)-thiosemicarbazide
-
Key words:
- quinoline
- / thiosemicarbazone
- / complex
- / crystal structure
- / DNA interaction
-
In the past few decades, thiosemicarbazones (TSCs) and their metal complexes have been a focus of chemists and biologists because of their wide range of pharmacological effects, such as antibacterial, antiviral, antifungal, and, most intriguingly, antitumor activity[1]. It is noted that the presence of a heterocyclic ring in the synthesized TSCs plays a major role in extending their pharmacological properties[1]. As a result, the transition metal complexes of TSCs derived from 2-acylpyridine/2-acylpyrazine have been exten-sively investigated as potential anticancer agents[1-11]. However, the studies on the complexes with TSCs bearing condensed heterocycles are relatively few [12-14].
On the other hand, the biological activities of TSCs metals depend on not only the structures of the ligands but also the metal centers[6-8]. In most case of TSCs, metal-ligand synergism could occur[1]. Recently, it has been demonstrated that several transition metal complexes with (quinolin-8-yl) methylene)-thiosemicarbazide have been reported to be potential antitumor agents[12-14]. As the continuation of our work on metal-TSCs[14], four transition metal complexes with 4-methyl-1-((quinolin-8-yl) methylene)-thiosemicarbazide (HL, Scheme 1) have been synthesized and structural determined by single-crystal X-ray diffraction. In addition, the interactions between the compounds and ct-DNA have been studied by ethidium bromide (EB) fluorescence probe.
1 Experimental
1.1 Materials and measurements
Solvents and starting materials for syntheses were purchased commercially and used as received. Elemental analyses were carried out on an Elemental Vario EL analyzer. 1H NMR spectra of HL was acquired with Bruker AV400 NMR instrument in DMSO-d6 solution with TMS as internal standard. The IR spectra (ν=4 000~400 cm-1) were determined by the KBr pressed disc method on a Bruker V70 FTIR spectrophotometer. The interactions between the compounds and ct-DNA are measured using literature method[14] via emission spectra on a Varian CARY Eclipse spectrophotometer.
1.2 Preparations of the ligand and complexes 1~4
The TSC ligand HL was produced according to the literature method, while using 4-methylthiosemicabazide instead of thiosemicabazide[13-14]. Anal. Calcd. For C12H12N4S (%): C: 58.99; H: 4.95; N: 22.93. Found (%): C: 59.12; H: 5.14; N: 22.86. FTIR (cm-1): ν(C=N)1 598, ν(C=N)quinoline1 545, ν(C=S) 822. 1H NMR (400 MHz) : δ 11.80 (s, 1H, NH), 9.75~9.77 (q, 1H, NH), 8.95~8.97(1H)/8.47~8.88(2H)/8.09~8.17(1H)/7.68~7.75(2H) for quinoline-H, 8.22 (s, 1H, CH =N), 3.00~3.04 (d, 3H, CH3).
The complexes 1, 2 and 4 were generated by reaction of HL (5 mmol) with equimolar of Ni (NO3)2, Zn (NO3)2 and CuCl2 in methanol (10 mL) solution, respectively, while using Cd (NO3)2 in ethanol (10 mL) solution for the synthesis of complex 3. Crystals of 1~4 suitable for X-ray diffraction analysis were obtained by evaporating the corresponding reaction solutions at room temperature.
1: brown rods. Anal. Calcd. For C26H30N8O2S2Ni (%): C: 51.24; H: 4.96; N: 18.39. Found (%): C: 51.32; H: 5.14; N: 18.26. FTIR (cm-1): ν(C=N)1 570, ν(N=C)quinoline1 482, ν(C-S)758.
2: yellow blocks. Anal. Calcd. for C25H26N8OS2Zn (%): C: 51.41; H: 4.49; N: 19.19. Found (%): C: 51.33; H: 4.29; N: 19.28. FTIR (cm-1): ν(C=N)1 571, ν(N=C)quinoline1 487, ν(C-S)755.
3: yellow blocks. Anal. Calcd. for C26H28N8OS2Cd (%): C: 48.41; H: 4.37; N: 17.37. Found (%): C: 48.44; H: 4.29; N: 17.45. FTIR (cm-1): ν(C=N)1 568, ν(N=C)quinoline1 495, ν(C-S)759.
4: green blocks. Anal. Calcd. for C24H24N8Cl4S2Cu2 (%): C: 38.05; H: 3.19; N: 14.79. Found (%): C: 38.18; H: 3.29; N: 14.68. FTIR (cm-1): ν(C=N)1 575, ν(N=C)quinoline1 502, ν(C=S)780.
1.3.1 X-ray crystallography
The X-ray diffraction measurement for complexes 1~4 was performed on a Bruker SMART APEX Ⅱ CCD diffractometer equipped with a graphite monochromatized Mo Kα radiation (λ=0.071 073 nm) by using φ-ω scan mode. Semi-empirical absorption correction was applied to the intensity data using the SADABS program[15]. The structures were solved by direct methods and refined by full matrix least-square on F2 using the SHELXTL-97 program[16]. All non-hydrogen atoms were refined anisotropically. All H atoms were positioned geometrically and refined using a riding model. SQUEEZE procedure was applied to deal with the existence of the voids of complex 3. Details of the crystal parameters, data collection and refinements for complexes 1~4 are summarized in Table 1.
 Table 1. Selected crystallographic data for complexes 1~4
Table 1. Selected crystallographic data for complexes 1~4CCDC: 1518183, 1; 1518184, 2; 1518185, 4; 1518186, 4.
2 Results and discussion
2.1 Crystal structures description
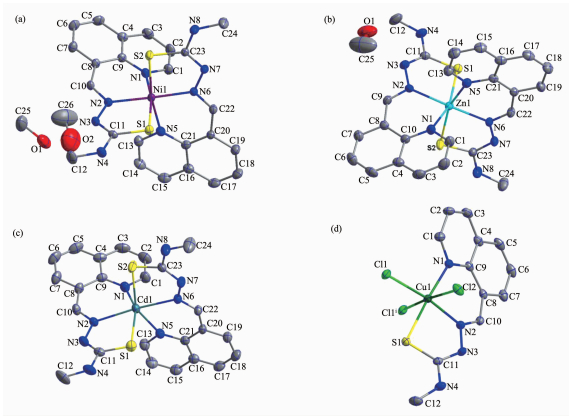
The diamond drawings of the asymmetric unit of 1~4 are shown in Fig. 1. Selected bond distances and angles are listed in Table 2. Hydrogen bonds information is in Table 3. The lengths of C-S bond are in the range of 0.169 0(5) and 0.173 3(5) nm in complexes 1~3, while that in complex 4 being 0.167 5(3) nm. This fact indicates that the ligand HL has thiolated and deprotonated in complexes 1~3[14], whilst acts as neutral ligand in complex 4.
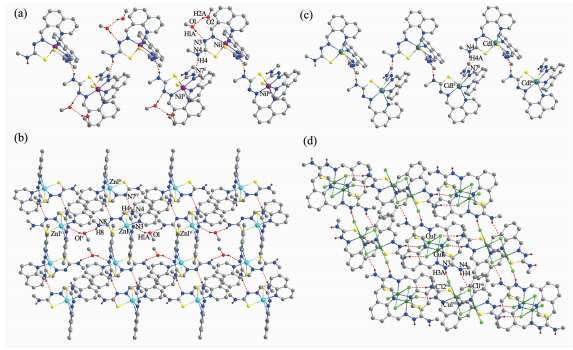
1 Nil-Nl 0.2l4 5(3) Nil-N2 0.205 6(3) Nil-N5 0.2l5 2(3) Nil-N6 0.205 0(3) Nil-Sl 0.238 34(l0) Nil-S2 0.239 0l (ll) N6-Nil-N2 l70.53(l0) Nl-Nil-N5 84.05(l0) N6-Nil-S2 8l.74(8) N6-Nil-Nl 98.85(l0) N6-Nil-Sl 9l.97(8) N2-Nil-S2 9l.70(8) N2-Nil-Nl 87.90(l0) N2-Nil-Sl 82.0l (8) Nl-Nil-S2 89.8l (8) N6-Nil-N5 88.79(l0) Nl-Nil-Sl l67.95(8) N5-Nil-S2 l67.8l (8) N2-Nil-N5 98.56(ll) N5-Nil-Sl 90.93(8) Sl-Nil-S2 97.0l (4) 2 Znl-Nl 0.224 0(4) Znl-N2 0.2l6 0(4) Znl-N5 0.225 5(4) Znl-N6 0.2l4 3(3) Znl-Sl 0.238 88(l3) Znl-S2 0.240 92(l5) N6-Znl-N2 l7l.74(l4) Nl-Znl-N5 80.42(l4) N6-Znl-S2 79.78(l0) N6-Znl-Nl l04.83(l4) N6-Znl-Sl 93.39(l0) N2-Znl-S2 96.98(l0) N2-Znl-Nl 82.53(l4) N2-Znl-Sl 80.04(l0) Nl-Znl-S2 87.9l (l2) N6-Znl-N5 84.56(l3) Nl-Znl-Sl l59.4l (ll) N5-Znl-S2 l57.39(l0) N2-Znl-N5 l00.58(l3) N5-Znl-Sl 92.07(l0) Sl-Znl-S2 l04.97(6) 3 Cdl-Nl 0.240 7(7) Cdl-N2 0.237 l (6) Cdl-N5 0.243 7(6) Cdl-N6 0.235 6(6) Cdl-Sl 0.252 6(2) Cdl-S2 0.254 3(3) N6-Cdl-N2 l7l.2(2) Nl-Cdl-N5 80.3(2) N6-Cdl-S2 75.63(l7) N6-Cdl-Nl ll0.7(2) N6-Cdl-Sl 95.43(l7) N2-Cdl-S2 l04.77(l7) N2-Cdl-Nl 78.l (2) N2-Cdl-Sl 76.l6(l6) Nl-Cdl-S2 89.4(2) N6-Cdl-N5 78.5(2) Nl-Cdl-Sl l50.00(l8) N5-Cdl-S2 l46.60(l7) N2-Cdl-N5 l04.0(2) N5-Cdl-Sl 9l.24(l7) Sl-Cdl-S2 lll.80(ll) 4 Cul-Nl 0.202 2(3) Cul-N2 0.l94 4(3) Cul-Sl 0.227 32(l9) Cul-Cll 0.223 7(2) Cul-Cl2 0.284 9(2) Cul-Cllⅰ 0.298 9(2) N2-Cul-Nl 90.22(l2) Cll-Cul-Sl 87.9l (6) N2-Cul-Cllⅰ 87.l4(9) N2-Cul-Cll l7l.67(7) N2-Cul-Cl2 86.8l (9) Nl-Cul-Cllⅰ 84.98(l0) Nl-Cul-Cll 97.7l (9) Nl-Cul-Cl2 92.33(l0) Cll-Cul-Cllⅰ 90.99(7) N2-Cul-Sl 83.96(9) Cll-Cul-Cl2 95.38(7) Sl-Cul-Cllⅰ 89.48(8) Nl-Cul-Sl l72.l6(7) Sl-Cul-Cl2 92.58(9) Cl2-Cul-Cllⅰ l73.37(3) Symmetry codes: ⅰ-x+2, -y, -z+2 Table 2. Selected bond lengths (nm) and angles (°) in complexes 1~4D-H…A d(D-H)/nm d(H…A)/nm d(D…A) nm ∠D-H…A/(°) 1 N (4)-H (4)…N (7)ⅱ 0.086 0.2l7 0.296 0(4) l53.4 0(l)-H (lA)…N (3) 0.082 0.206 0.287 9(4) l72.6 0(2)-H (2A)…0(l) 0.082 0.2l6 0.288 3(8) l47.6 2 N (4)-H (4)…N (7)ⅵ 0.086 0.219 0.301 9(6) 160.7 N (8)-H (8)…0(1)ⅴ 0.086 0.223 0.303 3(6) 156.1 0(1)-H (1A)…N (3) 0.082 0.221 0.294 4(7) 149.4 3 N (4)-H (4)…N (7)ⅱ 0.086 0.228 0.312 5(4) 169.2 4 N (3)-H (3A)…Cl (2)ⅶ 0.086 0.216 0.297 9(3) 160.0 N (4)-H (4)…Cl (1)ⅶ 0.086 0.258 0.325 7(4) 135.9 Symmetry codes: ⅱ-x+1, y+1/2, -z+1/2; ⅳ-x, y+1/2, -z+1/2; ⅴ x, -y+1/2, z+1/2; ⅶ-x+1, y-1/2, -z+3/2 Table 3. Hydrogen bonds information in complexes 1~4As shown in Fig. 1a~1c, the structures of complexes 1~3 are quite similar, while with different crystal solvent molecules. The metal ion in each complex with a distorted octahedron geometry is surrounded by two anionic TSC ligands with N4S2 donor set. The distances of metal-N/S bonds (Table 2) were comparable with those found in the reported complexes with similar donor set[14]. In the crystals of complexes 1 and 3, intermolecular N-H…N hydrogen bonds link the complex molecules into one-dimentional chains along b axis (Fig. 2a and 2c). Furthermore, the intermolecular O-H···N and OH…O hydrogen bonds between the lattice methanol molecules and complex molecules are also present in 1 (Table 3). By contrast, in the crystal of 2, the intermolecular N-H…N, N-H…O and O-H…N hydrogen bonds link the complexes and lattice methanol molecules into extended 2D supramolecular network (Fig. 2b).
However, complex 4 is binuclear, and the asymmetric unit contains a half of the molecule (Fig. 1d). Each central Cu(Ⅱ) ion of the dimer is surrounded by one independent neutral ligand and three coordinated chloride anions, two of which act as μ2-bridges, thus giving a distorted octahedron geometry. It should be noted that the bond lengths of Cu1-Cl2 and Cu1-Cl1ⅰ (Symmetry codes: ⅰ-x+2, -y, -z+2) are 0.284 9(2) and 0.298 9(2) nm, respectively, which are much longer than that of the normal Cu-Cl bond (Cu1-Cl1 0.223 7(2) nm). In the solid state, the intermolecular N-H…Cl hydrogen bonds are helpful to construct an extended 2D supramolecular network (Fig. 2d).
2.2 IR spectra
The most useful for determining the mode of coordination of the TSC ligand in infrared spectral bands are the ν(N=C), ν(N=C, quinoline) and ν(S=C) vibrations. Such three bonds of the free TSC is found at 1 598, 1 545 and 822 cm-1, while they shifts to lower frequency in complexes 1~4, clearly indicating the coordination of imine nitrogen, quinoline nitrogen and sulfur atoms [13-14], which is in accordance with the X-ray diffraction analysis result.
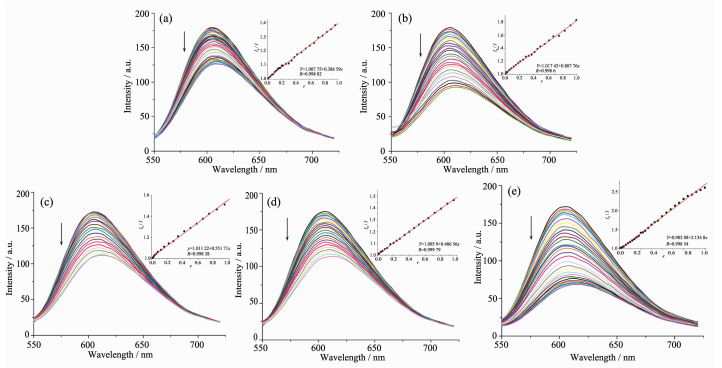
2.3 EB-DNA binding study by fluorescence spectrum
It is well known that EB can intercalate nonspecifically into DNA, which causes it to fluoresce strongly. Competitive binding of other drugs to DNA and EB will result in displacement of bound EB and thus decreasing the fluorescence intensity[17]. The effects of the ligand and complexes on the fluorescence spectra of EB-DNA system are presented in Fig. 3. The fluorescence intensities of EB bound to ct-DNA at about 600 nm show remarkable decreasing trends with the increasing concentration of the tested compounds, indicating that some EB molecules are released into solution after the exchange with the compounds. The quenching of EB bound to DNA by the compounds is in agreement with the linear Stern-Volmer equation: I0/I=1+Ksq r[18], where I0 and I represent the fluorescence intensities in the absence and presence of quencher, respectively, Ksq is the linear Stern-Volmer quenching constant, r is the ratio of the concentration of quencher and DNA. In the quenching plots of I0/I versus r, Ksq values are given by the slopes. The Ksq values are 0.808, 0.552, 0.487 and 2.315 for complexes 1~4, respectively, while that of the ligand HL is tested to be 0.387. The results indicate that interactions of the complexes with DNA are stronger than that of the ligand HL, probably due to the higher rigidity of the complexes, which is in accordance with the literature [14]. In addition, complex 4 has the highest quenching ability among the tested complexes, this could be explained from two aspects: (1) Cu(Ⅱ) plays a significant role in biological systems, and Cu(Ⅱ) complexes usually have higher biological activities than the other transition metal ones[19]; (2) the ligand HL in complex 4 is neutral, and thus the N-H bond at N2 position could act as an additional hydrogen bond donor, making it insert base pairs of DNA more easily than complexes 1~3.
-
-
[1]
Majid R, Hassan K. Coord. Chem. Rev., 2014, 280:203-253 doi: 10.1016/j.ccr.2014.06.007
-
[2]
Soares M A, Lessa J A, Mendes I C, et al. Bioorg. Med. Chem., 2012, 20:3396-3409 doi: 10.1016/j.bmc.2012.04.027
-
[3]
Qi J, Liang S, Gou Y, et al. Eur. J. Med. Chem., 2015, 96:360-368 doi: 10.1016/j.ejmech.2015.04.031
-
[4]
Li M X, Zhang L Z, Yang M, et al. Bioorg. Med. Chem. Lett., 2012, 22:2418-2433 doi: 10.1016/j.bmcl.2012.02.024
-
[5]
Li M X, Zhang L Z, Zhang D, et al. Eur. J. Med. Chem., 2011, 46:4383-4390 doi: 10.1016/j.ejmech.2011.07.009
-
[6]
Matesanz A I, Leitao I, Souza P. J. Inorg. Biochem., 2013, 125:26-31 doi: 10.1016/j.jinorgbio.2013.04.005
-
[7]
Matesanz A I, Souza P. Inorg. Chem. Commun., 2013, 27:5-8 doi: 10.1016/j.inoche.2012.10.022
-
[8]
Qi J, Su L, Gou Y, et al. Eur. J. Med. Chem., 2015, 96:360-368 doi: 10.1016/j.ejmech.2015.04.031
-
[9]
Jansson P J, Sharpe P C, Bernhardt P V, et al. J. Med. Chem., 2010, 53:5759-5769 doi: 10.1021/jm100561b
-
[10]
Bacher F, Enyedy E A, Nagy N V, et al. Inorg. Chem., 2013, 52:8895-8908 doi: 10.1021/ic401079w
-
[11]
Enyedy E A, Primik M F, Kowol C R, et al. Dalton Trans., 2011, 40:5895-5905 doi: 10.1039/c0dt01835j
-
[12]
Bourosh P N, Revenko M D, Stratulat E F, et al. Russ. J. Inorg.Chem., 2014, 59:545-557 doi: 10.1134/S0036023614060059
-
[13]
Revenko M D, Bourosh P N, Stratulat E F, et al. Russ. J. Inorg. Chem., 2010, 55:1387-1397 doi: 10.1134/S0036023610090093
-
[14]
毛盼东, 闫玲玲, 王文静, 等.无机化学学报, 2016, 32:555-561 https://www.researchgate.net/publication/301584710_Syntheses_Crystal_Structures_and_DNA-Binding_Properties_of_CoIICdII_Complexes_with_Quinoline_Thiosemicarbazone_LigandMAO Pan-Dong, YAN Ling-Ling, WANG Wen-Jing, et al. Chinese J. Inorg. Chem., 2016, 32:555-561 https://www.researchgate.net/publication/301584710_Syntheses_Crystal_Structures_and_DNA-Binding_Properties_of_CoIICdII_Complexes_with_Quinoline_Thiosemicarbazone_Ligand
-
[15]
Sheldrick G M. SADABS, University of Göttingen, Germany, 1996.
-
[16]
Sheldrick G M. SHELX-97, Program for the Solution and the Refinement of Crystal Structures, University of Göttingen, Germany, 1997.
-
[17]
Yang Z Y, Wang Y, Wang Y. Bioorg. Med. Chem. Lett., 2007, 17:2096-2101 doi: 10.1016/j.bmcl.2006.10.049
-
[18]
沈伟, 胡未极, 吴小勇, 等.无机化学学报, 2016, 32:1101-1110 http://www.researchgate.net/publication/279040924_Synthesis_Crystal_Structure_and_Biological_Activity_of_LanthanumIII_Coordination_Polymer_Constructed_with_p-Hydroxyphenylacetic_Acid_AnionSHEN Wei, HU Wei-Ji, WU Xiao-Yong, et al. Chinese J. Inorg. Chem., 2016, 32:1101-1110 http://www.researchgate.net/publication/279040924_Synthesis_Crystal_Structure_and_Biological_Activity_of_LanthanumIII_Coordination_Polymer_Constructed_with_p-Hydroxyphenylacetic_Acid_Anion
-
[19]
Nath M, Vats M, Roy P. Eur. J. Med. Chem., 2013, 59:310-321 doi: 10.1016/j.ejmech.2012.11.023
-
[1]
-
Figure 2 Chain-like structures along b axis formed via hydrogen bonds in complex 1(a) and 3(c); Extended 2D supramolecular structure in complex 2(b) and 4(d)
Hydrogen bonds shown in dashed line; H atoms for C-H bonds are omitted for clarity; Symmetry codes: ⅰ-x+2, -y, -z+2; ⅱ-x+1, y+1/2, -z+1/2; ⅲ-x+1, y-1/2, -z+1/2; ⅳ-x, y+1/2, -z+1/2; ⅴ x, -y+1/2, z+1/2; ⅵ x, -y+5/2, z-1/2; ⅶ-x+1, y-1/2, -z+3/2
Table 1. Selected crystallographic data for complexes 1~4

Table 2. Selected bond lengths (nm) and angles (°) in complexes 1~4
1 Nil-Nl 0.2l4 5(3) Nil-N2 0.205 6(3) Nil-N5 0.2l5 2(3) Nil-N6 0.205 0(3) Nil-Sl 0.238 34(l0) Nil-S2 0.239 0l (ll) N6-Nil-N2 l70.53(l0) Nl-Nil-N5 84.05(l0) N6-Nil-S2 8l.74(8) N6-Nil-Nl 98.85(l0) N6-Nil-Sl 9l.97(8) N2-Nil-S2 9l.70(8) N2-Nil-Nl 87.90(l0) N2-Nil-Sl 82.0l (8) Nl-Nil-S2 89.8l (8) N6-Nil-N5 88.79(l0) Nl-Nil-Sl l67.95(8) N5-Nil-S2 l67.8l (8) N2-Nil-N5 98.56(ll) N5-Nil-Sl 90.93(8) Sl-Nil-S2 97.0l (4) 2 Znl-Nl 0.224 0(4) Znl-N2 0.2l6 0(4) Znl-N5 0.225 5(4) Znl-N6 0.2l4 3(3) Znl-Sl 0.238 88(l3) Znl-S2 0.240 92(l5) N6-Znl-N2 l7l.74(l4) Nl-Znl-N5 80.42(l4) N6-Znl-S2 79.78(l0) N6-Znl-Nl l04.83(l4) N6-Znl-Sl 93.39(l0) N2-Znl-S2 96.98(l0) N2-Znl-Nl 82.53(l4) N2-Znl-Sl 80.04(l0) Nl-Znl-S2 87.9l (l2) N6-Znl-N5 84.56(l3) Nl-Znl-Sl l59.4l (ll) N5-Znl-S2 l57.39(l0) N2-Znl-N5 l00.58(l3) N5-Znl-Sl 92.07(l0) Sl-Znl-S2 l04.97(6) 3 Cdl-Nl 0.240 7(7) Cdl-N2 0.237 l (6) Cdl-N5 0.243 7(6) Cdl-N6 0.235 6(6) Cdl-Sl 0.252 6(2) Cdl-S2 0.254 3(3) N6-Cdl-N2 l7l.2(2) Nl-Cdl-N5 80.3(2) N6-Cdl-S2 75.63(l7) N6-Cdl-Nl ll0.7(2) N6-Cdl-Sl 95.43(l7) N2-Cdl-S2 l04.77(l7) N2-Cdl-Nl 78.l (2) N2-Cdl-Sl 76.l6(l6) Nl-Cdl-S2 89.4(2) N6-Cdl-N5 78.5(2) Nl-Cdl-Sl l50.00(l8) N5-Cdl-S2 l46.60(l7) N2-Cdl-N5 l04.0(2) N5-Cdl-Sl 9l.24(l7) Sl-Cdl-S2 lll.80(ll) 4 Cul-Nl 0.202 2(3) Cul-N2 0.l94 4(3) Cul-Sl 0.227 32(l9) Cul-Cll 0.223 7(2) Cul-Cl2 0.284 9(2) Cul-Cllⅰ 0.298 9(2) N2-Cul-Nl 90.22(l2) Cll-Cul-Sl 87.9l (6) N2-Cul-Cllⅰ 87.l4(9) N2-Cul-Cll l7l.67(7) N2-Cul-Cl2 86.8l (9) Nl-Cul-Cllⅰ 84.98(l0) Nl-Cul-Cll 97.7l (9) Nl-Cul-Cl2 92.33(l0) Cll-Cul-Cllⅰ 90.99(7) N2-Cul-Sl 83.96(9) Cll-Cul-Cl2 95.38(7) Sl-Cul-Cllⅰ 89.48(8) Nl-Cul-Sl l72.l6(7) Sl-Cul-Cl2 92.58(9) Cl2-Cul-Cllⅰ l73.37(3) Symmetry codes: ⅰ-x+2, -y, -z+2 Table 3. Hydrogen bonds information in complexes 1~4
D-H…A d(D-H)/nm d(H…A)/nm d(D…A) nm ∠D-H…A/(°) 1 N (4)-H (4)…N (7)ⅱ 0.086 0.2l7 0.296 0(4) l53.4 0(l)-H (lA)…N (3) 0.082 0.206 0.287 9(4) l72.6 0(2)-H (2A)…0(l) 0.082 0.2l6 0.288 3(8) l47.6 2 N (4)-H (4)…N (7)ⅵ 0.086 0.219 0.301 9(6) 160.7 N (8)-H (8)…0(1)ⅴ 0.086 0.223 0.303 3(6) 156.1 0(1)-H (1A)…N (3) 0.082 0.221 0.294 4(7) 149.4 3 N (4)-H (4)…N (7)ⅱ 0.086 0.228 0.312 5(4) 169.2 4 N (3)-H (3A)…Cl (2)ⅶ 0.086 0.216 0.297 9(3) 160.0 N (4)-H (4)…Cl (1)ⅶ 0.086 0.258 0.325 7(4) 135.9 Symmetry codes: ⅱ-x+1, y+1/2, -z+1/2; ⅳ-x, y+1/2, -z+1/2; ⅴ x, -y+1/2, z+1/2; ⅶ-x+1, y-1/2, -z+3/2 -

 扫一扫看文章
扫一扫看文章
计量
- PDF下载量: 3
- 文章访问数: 789
- HTML全文浏览量: 96


 下载:
下载:



 下载:
下载:

