
吡嗪缩氨基脲配体铜/锌配合物的晶体结构及荧光性质
English
Crystal Structures and Fluorescence Properties of Cu(Ⅱ)/Zn(Ⅱ) Complexes with a Semicarbazone Ligand Bearing Pyrazine Unit
-
Key words:
- semicarbazone
- / Cu(Ⅱ) complex
- / Zn(Ⅱ) complex
- / pyrazine
- / fluorescence
-
Schiff bases are an important class of ligands in coordination chemistry and have been paid much attention due to their extensive application in different fields[1-3]. Among them, thiosemicarbazones derived from acetyl-pyridine/pyrazine and their transition metal complexes have been extensively investigated as potential anticancer agents[3-12]. However, the study on the complexes of semicarbazones bearing pyrazine unit is relatively few[13-14].
On the other hand, both Cu(Ⅱ) and Zn(Ⅱ) ions are closely related to biochemistry, clinical diagnostics as well as environmental pollution[2, 14]. Our previous work has shown that the semicarbazone, namely, 1-(3-ethylpyrazin-2-yl) ethylidene-4-phenylsemicarbazide (HL) could act as a neutral ligand to form stable mononuclear complexes with Cu (NO3)2 and ZnCl2, respectively[14]. In addition, it has been demonstrated that the type of the metal salts could influence the structures of complexes efficiently[14]. Moreover, the polynuclear metal complexes are highly desired, because they usually possess higher anticancer activities than the mononuclear ones[2, 15]. Therefore, as a continuation of our work, two binu-clear complexes, [Cu2(L)2Br2]·CH3OH (1) and [Zn2(L)2(CH3COO)2]·2CH3OH (2) have been synthesized and structural determined by single-crystal X-ray diffrac-tion. The fluorescence properties of both complexes in methanol solution were also investigated.
1 Experimental
1.1 Materials and measurements
Solvents and starting materials for synthesis were purchased commercially and used as received. The ligand HL was prepared according to the literature method[14]. Elemental analysis was carried out on an Elemental Vario EL analyzer. The IR spectra (ν=4 000~400 cm-1) were determined by the KBr pressed disc method on a Bruker V70 FTIR spectrophotometer. The UV spectra were recorded on a Purkinje General TU-1800 spectrophotometer. Fluorescence spectra were determined on a Varian CARY Eclipse spectrophotometer, in the measurements of emission and excitation spectra the pass width is 5 nm.
1.2 Preparations of complexes 1 and 2
The complexes 1 and 2 were generated by reaction of the ligand HL (5 mmol) with equimolar of CuBr2 and Zn (OAc)2 in methanol solution (10 mL), respectively. Crystals suitable for X-ray diffraction analysis were obtained by evaporating the correspon-ding reaction solutions at room temperature.
1: Black blocks. Anal. Calcd. for C31H36N10O3 Br2Cu2 (%): C: 42.14; H: 4.11; N: 15.85. Found (%): C: 41.95; H: 4.22; N: 15.77. FTIR (cm-1): ν(N=C-O) 1625, ν(C=N) 1 580, ν(C=N)pyrazine 1 548.
2: Yellow blocks. Anal. Calcd. For C36H46N10O8Zn2 (%): C: 49.27; H: 5.28; N: 15.96. Found (%): C: 49.33; H: 5.38; N: 16.00. FTIR (cm-1): ν(N=C-O) 1 618, ν(C=N) 1 579, ν(C=N)pyrazine 1 552, νas (COO-) 1 569, νs (COO-) 1 436 and 1 413.
1.3.1 X-ray crystallography
The X-ray diffraction measurement for complexes 1 and 2 were performed on a Bruker SMART APEX Ⅱ CCD diffractometer equipped with a graphite monochromatized Mo Kα radiation (λ=0.071 073 nm) by using φ-ω scan mode. Semi-empirical absorption correction was applied to the intensity data using the SADABS program[16]. The structures were solved by direct methods and refined by full matrix least-square on F2 using the SHELXTL-97 program[17]. All non-hydrogen atoms were refined anisotropically. The H atoms of the disordered methanol molecule in 1 are not added. All the other H atoms were positioned geometrically and refined using a riding model. Details of the crystal parameters, data collection and refinements for complexes 1 and 2 are summarized in Table 1.
 Table 1. Crystal data and structure refinement for complexes 1 and 2
Table 1. Crystal data and structure refinement for complexes 1 and 2CCDC: 1516162, 1; 1516163, 2.
2 Results and discussion
2.1 Crystal structures description
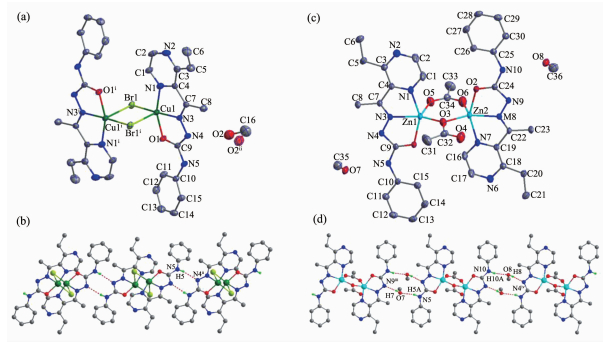
The diamond drawing of complexes 1 and 2 is shown in Fig. 1. Selected bond distances and angles are listed in Table 2. The lengths of C-O bond of the semicarbazone moiety are 0.125 8(4) nm (complex 1) or 0.125 6(4) and 0.125 3(4) nm (complex 2), which are longer than those of the reported neutral hydrazone or semicarbazones[13-14], indicating that the ligand HL has enolizated and deprotonated in both complexes. On the contrary, it acts as a neutral ligand in the literature complexes [Cu (HL)(H2O)(NO3)]NO3 and [Zn (HL) Cl2][14].
1 Br1-Cu1 0.235 90(6) Cu1-N1 0.200 4(3) Cu1-N3 0.192 4(3) Cu1-01 0.194 9(2) Cu1-Br1ⅰ 0.284 52(7) Br1-Cu1-Br1ⅰ 95.85(2) N3-Cu1-01 80.22(11) N3-Cu1-Br1 170.89(9) N3-Cu1-Br1ⅰ 93.04(9) N3-Cu1-N1 79.87(12) 01-Cu1-Br1 96.85(7) 01-Cu1-Br1ⅰ 95.90(8) 01-Cu1-N1 158.90(12) N1-Cu1-Br1 101.75(9) N1-Cu1-Br1ⅰ 92.05(9) 2 Zn1-01 0.206 9(3) Zn1-03 0.196 3(3) Zn1-05 0.198 6(3) Zn1-N1 0.215 5(3) Zn1-N3 0.202 7(3) Zn2-02 0.205 1(3) Zn2-03 0.196 9(3) Zn2-06 0.200 2(3) Zn2-N6 0.213 7(3) Zn2-N8 0.203 0(3) 05-Zn1-N3 109.51(12) N3-Zn1-01 76.72(11) 03-Zn1-05 105.72(13) 03-Zn1-01 99.83(11) 03-Zn1-N1 96.62(12) 03-Zn1-N3 144.25(12) 05-Zn1-01 105.33(13) 05-Zn1-N1 94.19(15) N3-Zn1-N1 75.16(11) 01-Zn1-N1 149.84(11) 03-Zn2-06 102.38(12) 03-Zn2-N8 148.00(12) 06-Zn2-02 93.54(13) 06-Zn2-N6 94.82(15) 06-Zn2-N8 109.51(12) N8-Zn2-02 76.82(11) N8-Zn2-N6 75.32(11) 03-Zn2-02 104.09(11) 03-Zn2-N6 100.00(11) 02-Zn2-N6 152.12(10) Symmetry codes: ⅰ-x+1, -y, -z+2 Table 2. Selected bond lengths (nm) and angles (°) in complexes 1 and 2As shown in Fig. 1a, complex 1 contains one centrosymmetric dimeric Cu(Ⅱ) molecule and one disordered methanol in the asymmetric unit. Two Cu atoms of the dimer were separated by 0.350 6 nm and doubly bridged by two bromide anions to form an ideal planar four-membered Cu2Br2 core. Each Cu(Ⅱ) ion is also coordinated by one L- anion with N2O donor set, giving a distorted square pyramid coordination geometry (τ=0.200)[13]. The basal plane of the square-pyramid is made up of N1, N3, O1 and Br1. The bond lengths from Cu(Ⅱ) center to these atoms are in the range of 0.192 4(3)~0.235 90(6) nm. The fifth coordination site is occupied by Br1ⅰ atom (Symmetry codes: ⅰ-x+1, -y, -z+2) located axially at 0.284 52(7) nm. In the solid state, the dimers were linked into a one-dimensional chain along c axis (Fig. 1b) by pairs of intermolecular N-H…N hydrogen bonds (Table 3).
D-H…A d(D-H)/nm d(H…A)/nm d(D…A) nm ∠DHA/(°) 1 N5-H5…N4ⅱ 0.086 0.217 0.300 2(4) 161.4 2 O7-H7…N9ⅲ 0.082 0.202 0.282 3(4) 167.8 O8-H8…N4ⅳ 0.082 0.205 0.286 5(4) 173.3 N5-H5A…O7 0.086 0.216 0.294 4(4) 151.8 N10-H10A…O8 0.086 0.22 0.298 2(4) 151.9 Symmetry codes: ⅱ-x+1, y, -z+3/2; ⅲ x, y+1, z; ⅳ x, y-1, z Table 3. Hydrogen bonds information in complexes 1 and 2Similarly, a discrete dimeric Zn(Ⅱ) molecule with Zn…Zn distance being 0.335 8 nm could also be observed in complex 2 (Fig. 1c). Each Zn(Ⅱ) ion adopts a distorted square pyramid coordination geometry (τ=0.093, 0.069 for Zn1 and Zn2, respectively)[13], and is surrounded by one L- and two acetate anions, one of which is bidentate bridge (μ-OCO), while the other one is oxygen bridge (μ-O). In the crystal, methanol molecules link the dimers into a one-dimensional chain along b axis (Fig. 1d) via intermolecular N-H…O and O-H…N hydrogen bonds.
2.2 IR spectra
The FTIR spectral region for both complexes is more or less similar due to the similar coordination modes of the ligand. The ν(C=O) of the free ligand at 1 705 cm-1 disappears in both complexes, meanwhile, new N=C-O stretching vibration absorption is observed at 1 625 and 1 618 cm-1 in complexes 1 and 2, respectively, revealing that the C=O in O=C-N moiety has enolizated and the oxygen atom coordinates to the metal ions in both complexes[13-14]. The ν(C=O), ν(C=N) and ν(C=N)pyrizine bands of the free ligand are at 1 609 and 1 595 cm-1, respectively. They shift to lower frequency values in the complexes, indicating that the imine N and pyrizine N atoms take part in the coordination[14]. In addition, the general pattern the IR spectroscopy of complex 1 supports the normal coordination of the μ-OCO and μ-O bridged acetate anions[18]. It is in accordance with the crystal structure study.
2.3 UV spectra
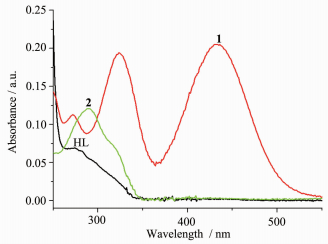
The UV spectra of complexes 1 and 2 in methanol solution (concentration: 1×10-5 mol· L-1) were measured at room temperature (Fig. 2). As our previous work shown, the spectra of HL features one main band located around 275 nm (ε=6 929 L·mol-1·cm-1) and a shoulder at 282 nm (ε=6 567 L·mol-1·cm-1), which could be assigned to characteristic π-π* transition of benzene and pyrazine units, respectively[14]. In complexes 1 and 2, the shoulder is absent and a combined band at 272 (ε=11 323 L·mol-1·cm-1) and 291 nm (ε=12 132 L·mol-1·cm-1) could be observed, respectively. The band at about 320 nm of complexes 1 (19 557 L·mol-1·cm-1) and 2 (ε=7 432 L·mol-1· cm-1, as shoulder) should be assigned to π→π* transition of imine bond[14, 18]. In addition, complex 1 exhibits another band at 434 nm (ε=20 511 L·mol-1·cm-1), corresponding to the ligand-to-metal charge transfer (LMCT)[19].
2.4 Fluorescence spectra
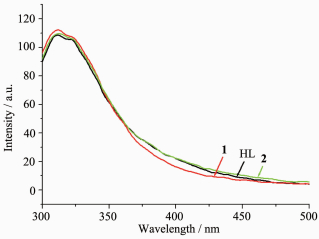
The fluorescence spectra of the ligand HL, complexes 1 and 2 have been studied in methanol solution (concentration: 1×10-5 mol· L-1) at room temperature. As shown in Fig. 3, the emission spectra of three compounds are quite similar, and each of them exhibits two indistinct peaks at 311 and 325 nm when excited at 285 nm. The emissions may be assigned to π*→π or π*→n transition of the intra-ligand[14, 19]. By contrast, the emission of the literature complex [Cu (HL)(H2O)(NO3)]NO3 has slightly red-shift (5 nm) when compared with that of HL[14]. Therefore, it may be roughly concluded that the type of the anions may influence not only the structure, but also the property of the complexes.
-
-
[1]
Dehghani-Firouzabadi A A, Sobhani M, Notash B. Polyhedron, 2016, 119:49-54 doi: 10.1016/j.poly.2016.08.021
-
[2]
Chang H Q, Jia L, Xu Z Q, et al. Inorg. Chem.Commun., 2015, 57:8-10 doi: 10.1016/j.inoche.2015.04.010
-
[3]
Majid Rezaeivala, Hassan Keypour. Coord. Chem. Rev., 2014, 280:203-253 doi: 10.1016/j.ccr.2014.06.007
-
[4]
Qi J, Liang S, Gou Y, et al. Eur. J. Med. Chem., 2015, 96:360-368 doi: 10.1016/j.ejmech.2015.04.031
-
[5]
Li M X, Zhang L Z, Yang M, et al. Bioorg. Med. Chem. Lett., 2012, 22:2418-2433 doi: 10.1016/j.bmcl.2012.02.024
-
[6]
Li M X, Zhang L Z, Zhang D, et al. Eur. J. Med. Chem., 2011, 46:4383-4390 doi: 10.1016/j.ejmech.2011.07.009
-
[7]
Matesanz A I, Leitao I, Souza P. J. Inorg. Biochem., 2013, 125:26-31 doi: 10.1016/j.jinorgbio.2013.04.005
-
[8]
Matesanz A I, Souza P. Inorg. Chem.Commun., 2013, 27:5-8 doi: 10.1016/j.inoche.2012.10.022
-
[9]
Kalinowski D S, Yu Y, Sharpe P C, et al. J. Med. Chem., 2007, 50:3716-3729 doi: 10.1021/jm070445z
-
[10]
Jansson P J, Sharpe P C, Bernhardt P V, et al. J. Med. Chem., 2010, 53:5759-5769 doi: 10.1021/jm100561b
-
[11]
Bacher F, Enyedy E A, Nagy N V, et al. Inorg. Chem., 2013, 52:8895-8908 doi: 10.1021/ic401079w
-
[12]
Enyedy E A, Primik M F, Kowol C R, et al. Dalton Trans., 2011, 40:5895-5905 doi: 10.1039/c0dt01835j
-
[13]
毛盼东, 韩学峰, 吴伟娜, 等.无机化学学报, 2016, 32:161-166 http://www.en.cnki.com.cn/Article_en/CJFDTotal-WJHX201601023.htmMAO Pan-Dong, HAN Xue-Feng, WU Wei-Na, et al. Chinese J. Inorg. Chem., 2016, 32:161-166 http://www.en.cnki.com.cn/Article_en/CJFDTotal-WJHX201601023.htm
-
[14]
毛献杰, 周利华, 伏思连, 等.无机化学学报, 2017, 33:163-168 http://www.ncbi.nlm.nih.gov/pubmed/17564456MAO Xian-Jie, ZHOU Li-Hua, FU Si-Lian, et al. Chinese J. Inorg. Chem., 2017, 33:163-168 http://www.ncbi.nlm.nih.gov/pubmed/17564456
-
[15]
Ma X Q, Zhu T F, Wu W N, et al. Indian J. Chem., 2016, 55A:1305-1313
-
[16]
Sheldrick G M. SADABS, University of Göttingen, Germany, 1996.
-
[17]
Sheldrick G M. SHELX-97, Program for the Solution and the Refinement of Crystal Structures, University of Göttingen, Germany, 1997.
-
[18]
Nakamoto K. Infrared and Raman Spectra of Inorganic and Coordination Compounds. 4th Ed. New York: Wiley, 1986: 257
-
[19]
王秀丽, 隋芳芳, 林宏艳, 等.无机化学学报, 2014, 30:2626-2634 https://www.researchgate.net/publication/279035432_Three_CuIICoII_Coordination_Polymers_Constructed_From_a_Flexible_Bis-pyridyl-bis-amide_and_Two_Different_Polycarboxylates_Assembly_Structure_and_PropertiesWANG Xiu-Li, SUI Fang-Fang, LIN Hong-Yan, et al. Chinese J. Inorg. Chem., 2014, 30:2626-2634 https://www.researchgate.net/publication/279035432_Three_CuIICoII_Coordination_Polymers_Constructed_From_a_Flexible_Bis-pyridyl-bis-amide_and_Two_Different_Polycarboxylates_Assembly_Structure_and_Properties
-
[1]
-
Figure 1 Diamond drawing of 1 (a) and 2 (b) with 30% thermal ellipsoids, extend chain-like structures along c axis in complex 1(c) and along b axis in complex 2 (d)
H atoms are omitted for clarity in (a) and (b); Disordered methanol are omitted in (c); Symmetry codes: ⅰ-x+1, -y, -z+2; ⅱ-x+1, y, -z+3/2; ⅲ x, y+1, z; ⅳ x, y-1, z
Table 1. Crystal data and structure refinement for complexes 1 and 2

Table 2. Selected bond lengths (nm) and angles (°) in complexes 1 and 2
1 Br1-Cu1 0.235 90(6) Cu1-N1 0.200 4(3) Cu1-N3 0.192 4(3) Cu1-01 0.194 9(2) Cu1-Br1ⅰ 0.284 52(7) Br1-Cu1-Br1ⅰ 95.85(2) N3-Cu1-01 80.22(11) N3-Cu1-Br1 170.89(9) N3-Cu1-Br1ⅰ 93.04(9) N3-Cu1-N1 79.87(12) 01-Cu1-Br1 96.85(7) 01-Cu1-Br1ⅰ 95.90(8) 01-Cu1-N1 158.90(12) N1-Cu1-Br1 101.75(9) N1-Cu1-Br1ⅰ 92.05(9) 2 Zn1-01 0.206 9(3) Zn1-03 0.196 3(3) Zn1-05 0.198 6(3) Zn1-N1 0.215 5(3) Zn1-N3 0.202 7(3) Zn2-02 0.205 1(3) Zn2-03 0.196 9(3) Zn2-06 0.200 2(3) Zn2-N6 0.213 7(3) Zn2-N8 0.203 0(3) 05-Zn1-N3 109.51(12) N3-Zn1-01 76.72(11) 03-Zn1-05 105.72(13) 03-Zn1-01 99.83(11) 03-Zn1-N1 96.62(12) 03-Zn1-N3 144.25(12) 05-Zn1-01 105.33(13) 05-Zn1-N1 94.19(15) N3-Zn1-N1 75.16(11) 01-Zn1-N1 149.84(11) 03-Zn2-06 102.38(12) 03-Zn2-N8 148.00(12) 06-Zn2-02 93.54(13) 06-Zn2-N6 94.82(15) 06-Zn2-N8 109.51(12) N8-Zn2-02 76.82(11) N8-Zn2-N6 75.32(11) 03-Zn2-02 104.09(11) 03-Zn2-N6 100.00(11) 02-Zn2-N6 152.12(10) Symmetry codes: ⅰ-x+1, -y, -z+2 Table 3. Hydrogen bonds information in complexes 1 and 2
D-H…A d(D-H)/nm d(H…A)/nm d(D…A) nm ∠DHA/(°) 1 N5-H5…N4ⅱ 0.086 0.217 0.300 2(4) 161.4 2 O7-H7…N9ⅲ 0.082 0.202 0.282 3(4) 167.8 O8-H8…N4ⅳ 0.082 0.205 0.286 5(4) 173.3 N5-H5A…O7 0.086 0.216 0.294 4(4) 151.8 N10-H10A…O8 0.086 0.22 0.298 2(4) 151.9 Symmetry codes: ⅱ-x+1, y, -z+3/2; ⅲ x, y+1, z; ⅳ x, y-1, z -

 扫一扫看文章
扫一扫看文章
计量
- PDF下载量: 1
- 文章访问数: 887
- HTML全文浏览量: 70


 下载:
下载:


 下载:
下载:

