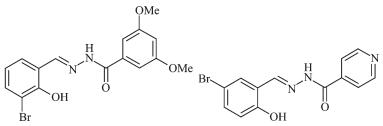
 Figure Scheme 1.
Ligands H2L1 (left) and H2L2 (right)
Figure Scheme 1.
Ligands H2L1 (left) and H2L2 (right)

酰腙配体双氧基钼(Ⅵ)配合物的合成、晶体结构和催化氧化性能
English
Syntheses, Crystal Structures and Catalytic Oxidation of Dioxomolybdenum(Ⅵ)Complexes with Hydrazone Ligands
-
Key words:
- hydrazone
- / molybdenum complex
- / crystal structure
- / catalytic property
-
0 Introduction
The mechanism of molybdenum oxotransferase has been extensively investigated for a long time. Molybdenum complexes are known for considerable use in organic chemistry, in particular for the various oxidations of organic compounds[1-5]. Dioxomolybdenum complexes have been intensively investigated as oxidation catalysts for variety of organic substrates, particularly for sulfoxidation and epoxidation of olefins[6-9]. The synthesis, characterization and reactivity studies of a number of dioxomolybdenum complexes with Schiff bases have been reported[10-13]. Some of the complexes have shown to possess oxygen atom transfer properties as they were found to oxidize thiols, hydrazine, polyketones and tertiary phosphines[14-15]. The catalytic ability of dioxomolybdenum(Ⅵ) complexes with benzohydrazone ligands toward the oxidation of sulfides have received satisfactory results[16-17]. However, the number of documented dioxomolybdenum(Ⅵ) complexes catalyzing the peroxidic oxidation of sulfides is still very limited. The rarely reports on such complexes are the preparation and structural characterization[18-21]. In the present paper, two new dioxomolybdenum(Ⅵ) complexes, [MoO2L1(MeOH)] (1) and [MoO2L2(MeOH)] (2) (Scheme 1; H2L1=N'-(3-bromo-2-hydroxybenzyli-dene)-3, 5-dimethoxybenzohydrazide for complex 1, H2L2=N'-(5-bromo-2-hydroxybenzylidene)isonicotinoh-ydrazide for complex 2), were prepared and studied for their structures and catalytic properties.
1 Experimental
1.1 Materials and methods
All chemicals and solvents were of analytical reagent grade, and purchased from Beijing Chemical Reagent Company. The ligands H2L1 and H2L2 were prepared according to the literature method[22]. Micro-analyses (C, H, N) were performed using a Perkin-Elmer 2400 elemental analyzer. Infrared spectra were carried out using the JASCO FT-IR model 420 spectrophotometer with KBr disk in the region of 4 000~400 cm-1. GC analyses were performed on a Shimadzu GC-2014 gas chromatograph.
1.2 Syntheses of the complexes
A hot methanol solution (15 mL) of MoO2(acac)2 (0.33 g, 1 mmol) was added to a hot methanol solution (15 mL) of H2L1 (0.38 g, 1 mmol) or H2L2 (0.31 g, 1 mmol). The mixture was refluxed for 30 minutes, and then cooled to room temperature, to give an orange solution. Single crystals suitable for X-ray diffraction were formed by slow evaporation of the solution containing the complex in air for a few days.
[MoO2L1(MeOH)] (1).Yield: 71%. Anal. Calcd. for C17H17BrMoN2O7(%): C, 38.0; H, 3.2; N, 5.2. Found(%): C, 37.8; H, 3.1; N, 5.3. IR data (KBr, cm-1): 3 453w, 1 601m, 1 441s, 1 358m, 1 164s, 1 073s, 949m, 859s, 740w, 540s, 463w. UV-Vis data (acetonitrile, λ / nm): 300 (15 510 L·moL-1·cm-1), 395 (3 020 L·moL-1·cm-1). ΛM (10-3 mol·L-1 in acetonitrile): 23 Ω-1·cm2·moL-1.
[MoO2L2(MeOH)] (2). Yield: 65%. Anal. Calcd. for C14H12BrMoN3O5(%): C, 35.2; H, 2.5; N, 8.8. Found(%): C, 35.3; H, 2.6; N, 8.7. IR data (KBr, cm-1): 3 455w, 1 629m, 1 434s, 1 150s, 1 081s, 956m, 851s, 533s. UV-Vis data (acetonitrile, λ / nm): 275 (14 320 L·moL-1·cm-1), 405 (3 900 L·moL-1·cm-1). ΛM (10-3 mol·L-1 in acetonitrile): 35 Ω-1·cm2·moL-1.
1.3 X-ray structure determination
Single crystal X-ray diffraction measurement was performed using a Bruker Smart 1000 CCD diffracto-meter with graphite monochromated Mo Kα radiation (λ=0.071 073 nm) using the φ-ω scan technique. The positions of non-hydrogen atoms were located with direct methods. Subsequent Fourier syntheses were used to locate the remaining non-hydrogen atoms. All non-hydrogen atoms were refined anisotropically. The methanol hydrogen atoms were located from difference Fourier maps and refined isotropically, with O-H distances restrained to 0.085(1) nm. The remaining hydrogen atoms were placed in calculated positions and constrained to ride on their parent atoms. The analysis was performed with the SHELXS-97 and SHELXL-97 program[23-24]. The crystallographic data for the complex are summarized in Table 1. Selected bond lengths and angles are given in Table 2.
1 2 Formula C17H17BrMoN2O7 C14H12BrMoN3O5 Formula weight 537.18 478.12 Crystal system Monoclinic Triclinic Space group P21/n P1 a/nm 0.848 98(3) 0.655 05(9) b/nm 2.275 4(1) 1.076 63(7) c/nm 1.069 82(5) 1.303 3(1) α/(°) 90 67.383(2) β/(°) 109.108(1) 84.264(1) γ/(°) 90 76.195(2) V/nm3 1.952 8(1) 0.823 9(1) Z 4 2 μ/mm-1 (Mo Kα) 2.760 3.250 Dc/(g·cm-3) 1.827 1.927 Independent reflections 3 576 3 006 Observed reflections [I≥2σ(I)] 3 105 1 975 Parameters 259 221 Restraints 1 1 Rint 0.024 5 0.068 1 Final R indices [I≥2σ(I)] R1=0.036 7, wR2=0.084 0 R1=0.071 3, wR2=0.151 0 R indices (all data) R1=0.045 9, wR2=0.089 8 R1=0.108 8, wR2=0.181 3 1 Mo1-O1 0.192 4(3) Mo1-O2 0.201 7(3) Mo1-O5 0.170 0(3) Mo1-O6 0.231 0(3) Mo1-O7 0.168 5(3) Mo1-N1 0.224 4(3) O1-Mo1-O2 148.27(11) O1-Mo1-N1 80.48(11) O5-Mo1-N1 160.37(13) O2-Mo1-N1 71.43(10) O5-Mo1-O1 104.46(13) O5-Mo1-O2 97.70(13) O7-Mo1-O1 99.10(16) O7-Mo1-O2 96.89(15) O7-Mo1-O5 104.65(15) O7-Mo1-N1 93.08(13) O7-Mo1-O6 171.96(12) O5-Mo1-O6 82.89(12) O1-Mo1-O6 81.42(12) O2-Mo1-O6 79.09(11) N1-Mo1-O6 79.06(10) 2 Mo1-O1 0.192 1(5) Mo1-O2 0.201 6(5) Mo1-O3 0.233 0(6) Mo1-O4 0.170 1(5) Mo1-O5 0.168 8(6) Mo1-N1 0.224 2(6) O1-Mo1-O2 149.5(2) O1-Mo1-N1 82.1(2) O2-Mo1-N1 71.7(2) O4-Mo1-O1 102.6(3) O4-Mo1-N1 162.2(3) O4-Mo1-O2 98.2(3) O5-Mo1-O1 99.2(3) O5-Mo1-O2 96.5(3) O5-Mo1-O3 167.8(3) O5-Mo1-O4 105.2(3) O5-Mo1-N1 90.7(3) O4-Mo1-O3 86.5(3) O1-Mo1-O3 81.2(2) O2-Mo1-O3 78.1(2) N1-Mo1-O3 77.2(2) CCDC: 1476621, 1; 1476622, 2.
2 Results and discussion
2.1 Synthesis
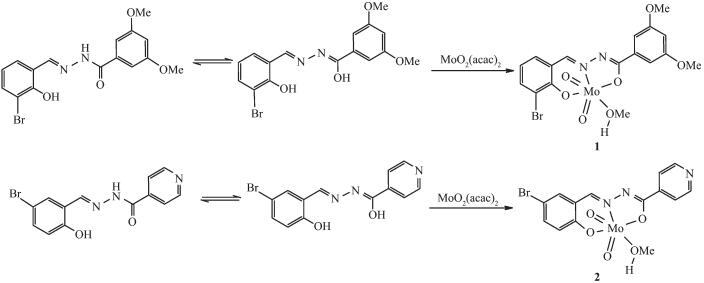
The hydrazone ligands H2L1 and H2L2 were readily prepared by the condensation reactions of 3-bromosalicylaldehyde with 3, 5-dimethoxybenzohydra-zide, and 5-bromosalicylaldehyde with isonicotinohy-drazide, respectively, in methanol. The dioxomolybd-enum(Ⅵ) complexes were synthesized by refluxing hydrazone ligands and MoO2(acac)2 in methanol in a 1:1 molar ratio. The reaction progress (Scheme 2) is accompanied by a color change of the solution from colorless to orange. We have attempted to prepare and grow diffraction quality crystals from various solvents, however, good quality crystals were finally obtained from methanol. The chemical formulae of the complexes have been confirmed by elemental analyses, IR spectra, and X-ray single crystal structure determination.
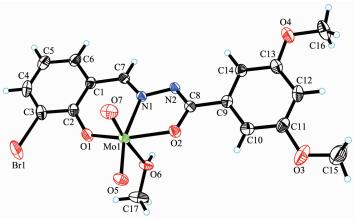
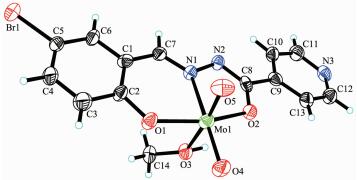
2.2 Structure description of the complexes
The molecular structures of complexes 1 and 2 are shown in Fig. 1 and 2, respectively. The coordination geometry around the Mo atom can be described as a slightly distorted octahedral geometry, with the phenolic O, imino N, and enolic O atoms of the hydrazone ligand, one oxo O atom defining the equatorial plane, and with one methanol O atom and the other oxo O atom occupying the axial positions. The hydrazone ligand coordinates to the Mo atom in a meridional fashion forming five-and six-membered chelate rings with bite angles of 71.43(10)° and 80.48(11)° for 1 and 71.7(2)° and 82.1(2)° for 2. The dihedral angles between the aromatic rings of the hydrazone ligands are 11.2(3)° for 1 and 14.1(3)° for 2. The displacements of the Mo atoms in complexes 1 and 2 from the equatorial mean planes toward the apical oxo atoms are 0.030 5(1) nm and 0.029 2(1) nm, respectively. The hydrazone ligands are coordinated in their dianionic form, which is evident from the N2-C8 and O2-C8 bond lengths in the hydrazone ligands. The abnormal bond values indicate the presence of the enolate form of the ligand amide groups. The Mo-O, Mo-N, and Mo=O bonds (Table 2) are within normal ranges and are similar to those observed in similar dioxomolybdenum(Ⅵ) complexes[18-21, 25-28].
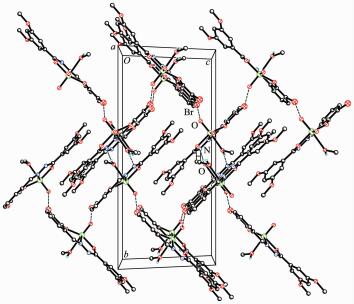
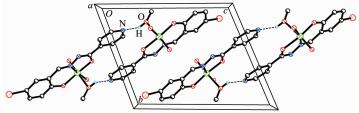
In the crystal structure of complex 1, adjacent two [MoO2L1] units are linked through two methanol ligands via hydrogen bonds (O6-H6 0.085(1) nm, H6…N2ⅰ 0.192(2) nm, O6…N2ⅰ 0.275 4(4) nm, O6-H6…N2ⅰ 170(6)°, Symmetry codes: ⅰ -x, -y, 1-z), to form a dimer. The dimers are further linked through weak Br…O interactions, to form a two-dimensional sheet (Fig. 3). In the crystal structure of complex 2, adjacent two [MoO2L2] units are linked through two methanol ligands via hydrogen bonds (O3-H3 0.085(1) nm, H3…N3ⅱ 0.196(7) nm, O3…N3ⅱ 0.267 7(9) nm, O3-H3…N3ⅱ 141(10)°, Symmetry codes: ⅱ 1-x, 1-y, -z), to form a dimer (Fig. 4).
2.3 IR spectra
The hydrazone ligands show stretching bands attributed to C=O, C=N, C-OH and NH at about 1 650, 1 610~1 620, 1 180, and 3 250 cm-1, respectively[29]. For the complexes, absence of the bands characteristic of the N-H and C=O groups indicates enolization of the hydrazone ligands and coordination through the deprotonated enolic-oxygen atom. The Mo=O stretching mode occur as single and medium bands at 949 cm-1 for 1 and 956 cm-1 for 2, assigned to the asymmetric stretching mode of the MoO2 moieties[29-30]. The strong bands indicative of the C=N groups of the complexes are located at 1 601 cm-1 for 1 and 1 629 cm-1 for 2[31].
Electronic spectra of the complexes recorded in acetonitrile solution display strong and medium absorption bands in the regions of 380~420 and 270~300 nm. These peaks are assigned as charge transfer transitions of the type N(pπ)-Mo(dπ) LMCT and O(pπ) -Mo(dπ) LMCT, respectively[22-23], as the ligand based orbitals are either N or O donor types. The slight changes of λmax values within each set of peaks may be due to the difference of electron donating capacity of the hydrazone ligands.
2.4 Catalytic oxidation
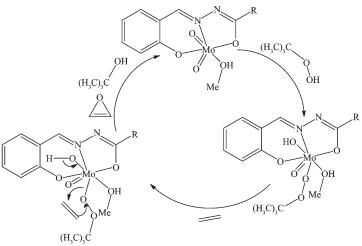
The catalytic experiment was carried out according to the literature method by using tert-butyl hydrogen peroxide (TBHP) as oxidant[9]. The results are listed in Table 3. Generally, excellent epoxide yields and selectivities were observed for all aliphatic and aromatic substrates. Oxidation of aromatic substrates gave the corresponding epoxides in 100% yields, while in the oxidation of aliphatic substrates, the conversion is less than 100%. Based on this consideration, it can be observed that the isolated double bonds are less reactive than the conjugated ones. The phenomenon is in good agreement with those reported previously[9]. There is no distinct difference for the catalytic property between the two complexes. The proposed catalytic mechanism of the catalytic reactions is shown in Scheme 3. First, TBHP was activated by coordination to the Mo atom and formation of hepta-coordinated molybdenum intermediate. Then a molecule of an olefin as a nucleophile attacks the electrophilic oxygen atom of the coordinated TBHP. As a result, the corresponding epoxides were formed, and TBHP was reduced to tert-butyl alcohol.
Substrate Product Conversionb (TON)c/% Selectivity/% 

1 93 (272) 100 2 91 (261) 100 

1 89 (258) 100 2 85 (263) 100 

1 96 (282) 100 2 93 (278) 100 

1 100 (303) 100 2 100 (291) 100 

1 100 (252) 100 2 100 (270) 100 

1 100 (311) 100 2 100 (294) 100 

1 100 (305) 100 2 100 (310) 100 

1 100 (245) 100 2 100 (257) 100 a ncatalyst:nsubstrate:nTBHP=1:300:1 000, Reactions were performed in mixture of CH3OH/CH2Cl2 (6:4, V/V, 1.5 mL); b GC conversion (%) was measured relative to the starting substrate after 1 h; c TON=nproduct/ncatalyst 3 Conclusions
Two new dioxomolydenum(Ⅵ) complexes with similar hydrazone ligands, N'-(3-bromo-2-hydroxyben-zylidene)-3, 5-dimethoxybenzohydrazide and N'-(5-bromo-2-hydroxybenzylidene)isonicotinohydrazide, have been prepared and structurally characterized by single crystal X-ray structure determination and infrared and electronic spectra. The hydrazone ligands coordinate to the Mo atoms through the phenolic O, imino N and ethanolic O atoms. Both complexes have efficient catalytic properties on the oxidation of various olefins to their corresponding epoxides.
-
-
[1]
Feng L S, Maass J S, Luck R L. Inorg. Chim. Acta, 2011, 373: 85-92 doi: 10.1016/j.ica.2011.03.060
-
[2]
Gamelas C A, Gomes A C, Bruno S M, et al. Dalton Trans., 2012, 41:3474-3484 doi: 10.1039/c2dt11751g
-
[3]
Bagherzadeh M, Ghazali-Esfahani S. New J. Chem., 2012, 36: 971-976 doi: 10.1039/c2nj21001k
-
[4]
Amarante T R, Neves P, Tome C, et al. Inorg. Chem., 2012, 51: 3666-3676 doi: 10.1021/ic202640a
-
[5]
Neuenschwander J, Meier E, Hermans I. Chem. Eur. J., 2012, 18:6776-6780 doi: 10.1002/chem.201200470
-
[6]
Krackl S, Company A, Enthaler S, et al. ChemCatChem, 2011, 3:1186-1192 doi: 10.1002/cctc.v3.7
-
[7]
Grover N, Kuhn F E. Curr. Org. Chem., 2012, 16:16-32 doi: 10.2174/138527212798993103
-
[8]
Jin N Y. J. Coord. Chem., 2012, 65:4013-4022 doi: 10.1080/00958972.2012.731049
-
[9]
Rayati S, Rafiee N, Wojtczak A. Inorg. Chim. Acta, 2012, 386: 27-35 doi: 10.1016/j.ica.2012.02.005
-
[10]
Gao S P. J. Coord. Chem., 2011, 64:2869-2877 doi: 10.1080/00958972.2011.608163
-
[11]
Tangestaninejad S, Moghadam M, Mirkhani V, et al. Catal. Commun., 2009, 10:853-858 doi: 10.1016/j.catcom.2008.12.010
-
[12]
El-Tabl A S, Issa R M, Morsi M A. Transition Met. Chem., 2004, 29:543-549 doi: 10.1023/B:TMCH.0000037524.95228.00
-
[13]
Grivani G, Tangestaninejad S, Halili A. Inorg. Chem. Commun., 2007, 10:914-917 doi: 10.1016/j.inoche.2007.01.016
-
[14]
Sheikhsoaie I, Rezaeffard A, Monadi N, et al. Polyhedron, 2009, 28:733-738 doi: 10.1016/j.poly.2008.12.044
-
[15]
Rao S N, Kathale N, Rao N N, et al. Inorg. Chim. Acta, 2007, 360:4010-4016 doi: 10.1016/j.ica.2007.05.035
-
[16]
Debel R, Buchholz A, Plass W. Z. Anorg. Allg. Chem., 2008, 634:2291-2298 doi: 10.1002/zaac.v634:12/13
-
[17]
Mancka M, Plass W. Inorg. Chem. Commun., 2007, 10:677-680 doi: 10.1016/j.inoche.2007.02.029
-
[18]
Vrdoljak V, Prugovecki B, Matkovic-Calogovic D, et al. Cryst. Growth Des., 2011, 11:1244-1252 doi: 10.1021/cg1014576
-
[19]
Zhai Y L, Xu X X, Wang X. Polyhedron, 1992, 11:415-418 doi: 10.1016/S0277-5387(00)83195-2
-
[20]
Sergienko V S, Abramenko V L, Minacheva L K, et al. Koord. Khim., 1993, 19:28-37
-
[21]
Lei Y, Fu C. Synth. React. Inorg. Met.-Org. Nano-Met. Chem., 2011, 41:704-709 doi: 10.1080/15533174.2011.568465
-
[22]
Xu W X, Li W H. Synth. React. Inorg. Met.-Org. Nano-Met. Chem., 2012, 42:160-164 doi: 10.1080/15533174.2011.609511
-
[23]
Sheldrick G M. SHELX-97, Program for Crystal Structure Solution and Refinement, University of Göttingen, Germany, 1997.
-
[24]
Sheldrick G M. SHELXTL Version 5, Siemens Industrial Automation Inc. , Madison, 1995.
-
[25]
Dinda R, Ghosh S, Falvello L R, et al. Polyhedron, 2006, 25: 2375-2382 doi: 10.1016/j.poly.2006.02.002
-
[26]
Dinda R, Sengupta S, Ghosh S, et al. J. Chem. Soc. Dalton Trans., 2002, 23:4434-4439 http://pubs.rsc.org/en/content/articlelanding/2002/dt/b207129k#!divAbstract
-
[27]
Gupta S, Barik A K, Pal S, et al. Polyhedron, 2007, 26:133-141 doi: 10.1016/j.poly.2006.08.001
-
[28]
Bansse W, Ludwig E, Schilde U, et al. Z. Anorg. Allg. Chem., 1995, 621:1275-1281 doi: 10.1002/(ISSN)1521-3749
-
[29]
Rao S N, Munshi K N, Rao N N, et al. Polyhedron, 1999, 18:2491-2497 doi: 10.1016/S0277-5387(99)00139-4
-
[30]
El-Medani S M, Aboaly M M, Abdalla H H, et al. Spectrosc. Lett., 2004, 37:619-632 doi: 10.1081/SL-200037610
-
[31]
Ngan N K, Lo K M, Wong C S R. Polyhedron, 2011, 30: 2922-2932 doi: 10.1016/j.poly.2011.08.038
-
[1]
-
Table 1. Crystal data for the complexes
1 2 Formula C17H17BrMoN2O7 C14H12BrMoN3O5 Formula weight 537.18 478.12 Crystal system Monoclinic Triclinic Space group P21/n P1 a/nm 0.848 98(3) 0.655 05(9) b/nm 2.275 4(1) 1.076 63(7) c/nm 1.069 82(5) 1.303 3(1) α/(°) 90 67.383(2) β/(°) 109.108(1) 84.264(1) γ/(°) 90 76.195(2) V/nm3 1.952 8(1) 0.823 9(1) Z 4 2 μ/mm-1 (Mo Kα) 2.760 3.250 Dc/(g·cm-3) 1.827 1.927 Independent reflections 3 576 3 006 Observed reflections [I≥2σ(I)] 3 105 1 975 Parameters 259 221 Restraints 1 1 Rint 0.024 5 0.068 1 Final R indices [I≥2σ(I)] R1=0.036 7, wR2=0.084 0 R1=0.071 3, wR2=0.151 0 R indices (all data) R1=0.045 9, wR2=0.089 8 R1=0.108 8, wR2=0.181 3 Table 2. Selected bond lengths (nm) and angles (°) for the complexes
1 Mo1-O1 0.192 4(3) Mo1-O2 0.201 7(3) Mo1-O5 0.170 0(3) Mo1-O6 0.231 0(3) Mo1-O7 0.168 5(3) Mo1-N1 0.224 4(3) O1-Mo1-O2 148.27(11) O1-Mo1-N1 80.48(11) O5-Mo1-N1 160.37(13) O2-Mo1-N1 71.43(10) O5-Mo1-O1 104.46(13) O5-Mo1-O2 97.70(13) O7-Mo1-O1 99.10(16) O7-Mo1-O2 96.89(15) O7-Mo1-O5 104.65(15) O7-Mo1-N1 93.08(13) O7-Mo1-O6 171.96(12) O5-Mo1-O6 82.89(12) O1-Mo1-O6 81.42(12) O2-Mo1-O6 79.09(11) N1-Mo1-O6 79.06(10) 2 Mo1-O1 0.192 1(5) Mo1-O2 0.201 6(5) Mo1-O3 0.233 0(6) Mo1-O4 0.170 1(5) Mo1-O5 0.168 8(6) Mo1-N1 0.224 2(6) O1-Mo1-O2 149.5(2) O1-Mo1-N1 82.1(2) O2-Mo1-N1 71.7(2) O4-Mo1-O1 102.6(3) O4-Mo1-N1 162.2(3) O4-Mo1-O2 98.2(3) O5-Mo1-O1 99.2(3) O5-Mo1-O2 96.5(3) O5-Mo1-O3 167.8(3) O5-Mo1-O4 105.2(3) O5-Mo1-N1 90.7(3) O4-Mo1-O3 86.5(3) O1-Mo1-O3 81.2(2) O2-Mo1-O3 78.1(2) N1-Mo1-O3 77.2(2) Table 3. Details of the catalytic oxidation of olefins catalyzed by the complexesa
Substrate Product Conversionb (TON)c/% Selectivity/% 

1 93 (272) 100 2 91 (261) 100 

1 89 (258) 100 2 85 (263) 100 

1 96 (282) 100 2 93 (278) 100 

1 100 (303) 100 2 100 (291) 100 

1 100 (252) 100 2 100 (270) 100 

1 100 (311) 100 2 100 (294) 100 

1 100 (305) 100 2 100 (310) 100 

1 100 (245) 100 2 100 (257) 100 a ncatalyst:nsubstrate:nTBHP=1:300:1 000, Reactions were performed in mixture of CH3OH/CH2Cl2 (6:4, V/V, 1.5 mL); b GC conversion (%) was measured relative to the starting substrate after 1 h; c TON=nproduct/ncatalyst -

 扫一扫看文章
扫一扫看文章
计量
- PDF下载量: 0
- 文章访问数: 810
- HTML全文浏览量: 113


 下载:
下载:






 下载:
下载:

