
两个菲咯啉铜配合物的合成、晶体结构和催化性能
English
Syntheses, Crystal Structures and Catalytic Performances of Two Cu Complexes with 1,10-Phenanthroline Ligand
-
Key words:
- Cu complexes
- / solvothermal synthesis
- / dimethyl carbonate
- / oxidative carbonylation
-
0 Introduction
Copper-haloid complexes have rich structural motifs, such as monomeric species[1], rhomboid Cu2×2 dimers[2], cubane or stepped-cubane Cu4×4 tetramers[3], Cu6×6 clusters[4], zigzag [CuX]n chains[5], double-stranded [Cu2×2]n ladders[6], [Cu6×6]n banded ribbons[7], and 2D [CuX]n layers[8]. Among them, monovalent copper compounds exhibit strong fluorescence, electrical con-ductivity, and catalytic properties[9], divalent copper compounds exhibit magnetic, biological and catalytic properties[10], and mixed-valent Cu(Ⅰ)-Cu(Ⅱ) copper compounds exhibit biological and electronic proper-ties[11]. For controlling the topologies and properties of these copper-haloid complexes, it is important to rationally select ligands, type of the anions, and reaction conditions. Hence, the design and assembly of copper-haloid complexes have attracted increasing interest as an expanding field[12].
Herein, a Cu(Ⅰ)-Cu(Ⅱ) mixed-valent Cu complex, [Cu(phen)2Br]2[Cu4Br6] (1) (phen=1,10-phenanthroline) was obtained from CuBr2 and phen in alcohol by auto-reduction, and the other Cu complex, [Cu(phen)2Br]Br·CH3OH (2), was synthesized from CuBr and phen in methanol by oxidation. Their structures were inve-stigated by infrared spectroscopy, thermal analysis, and X-ray diffraction (XRD) single-crystal structure analysis, and their catalytic activities were inves-tigated for the oxidative carbonylation of methanol.
1 Experimental
1.1 Materials and measurements
Methanol and anhydrous ethanol were freshly distilled prior to use. Other reagents were used as received without further purification. Elemental analyses were carried out on an elemental Vario EL analyzer. Diffraction intensity data were collected on a Bruker SMART APEX-Ⅱ CCD diffractometer. The thermal investigations were done on a Q50 therm-alanalyzer under a dynamic nitrogen environment with a heating rate of 10 ℃·min-1. Infrared spectra were recorded on a Nicolet 6700 FT-IR spectrophotometer in the form of KBr pellets.
1.2 Syntheses of the complexes
Synthesis of [Cu(phen)2Br]2[Cu4Br6] (1): First, 4 mmol (0.792 g) of 1,10-phenanthroline dihydrate in 15.0 mL of ethanol was added dropwise to a solution containing 4 mmol (0.893 g) of CuBr2 and 15.0 mL of methanol, followed by stirring for 30 min at 25 ℃ and filtered. Second, the residue and 25.0 mL of ethanol were added into a Parr Teflon-lined autoclave (50 mL) and heated at 150 ℃ for 3 days. After cooling down to room temperature, dark green crystals were obtained. Yield: 52.7% (based on Cu). Anal. Calcd. for C24H16Br4Cu3N4(%): C, 33.11; H, 1.85; N, 6.43; Found(%): C, 32.90; H, 1.85; N, 6.27. FT-IR (KBr, cm-1): 3 048 (w), 1 622 (m), 1 605 (w), 1 582 (w), 1 519 (s), 1 428 (s), 858 (s), 721 (s), 493 (m).
Synthesis of [Cu(phen)2Br]Br·CH3OH (2): First, CuBr (0.431 g, 3 mmol) was added into methanol (20 mL). Second, the mixture was refluxed for 6 h under oxygen, followed by the addition of 1,10-phenanthroline dihydrate (0.594 g, 3 mmol). Third, the solution was refluxed again for 8 h, followed by filtration. After the solution was cooled to room temperature, the filtrate was placed in a refrigerator for two weeks to afford green crystals. Yield: 53.1% (based on phen). Anal. Calcd. for C25H20Br2CuN4O(%): C, 48.76; H, 3.27; N, 9.10. Found(%): C, 49.09; H, 2.99; N, 9.38. FT-IR (KBr, cm-1): 3 030 (vw), 1 627 (m), 1 605 (w), 1 586 (m), 1 518 (s), 1 427 (s), 1 104 (m), 852 (s), 722 (s), 430 (m).
1.3 X-ray data collection and structure refinements
Reflection intensities of the two crystals were collected on a Bruker APEX-Ⅱ CCD diffractometer with Mo Kα radiation (λ=0.071 073 nm). Lp correction and a ψ empirical absorption correction were made for intensity data. The structures of 1 and 2 were solved by direct methods, and further refined by the full-matrix least-squares method on F2 with anisotropic displacement parameters for all non-hydrogen atoms[13-16]. Hydrogen atoms associated with carbon atoms were geometrically generated, and the remaining hydrogen atoms were located from the difference Fourier maps. Hydrogen atoms were further refined isotropically using the riding model. In the final refinement, four copper atoms in 1 were found to be disordered and were represented by two sets of atomic positions (Cu(2), Cu(3), Cu(4), Cu(5) and Cu(2A), Cu(3A), Cu(4A), Cu(5A)). The occupancy for each of them was 0.5. The solvate methanol in 2 was orientational disorder, and the occupancies for all the atoms were 0.25. Table 1 summarizes the details of crystallographic data and structure refinement for two complexes.
Complex 1 2 Empirical formula C24H16Br4Cu3N4 C25H20Br2CuN4O Formula weight 870.67 615.81 Crystal system Triclinic Monoclinic Space group p1 C2/c a/nm 1.033 94(19) 1.428 0(4) b/nm 1.129 6(2) 1.494 4(4) c/nm 1.243 9(2) 1.243 6(4) α/(°) 78.194(4) 90.00 β/(°) 84.108(4) 106.638(5) γ/(°) 82.581(4) 90.00 V/nm3 1.405 8(5) 2.542 8(13) Z 2 4 Crystal size/mm 0.20×0.20×0.18 0.20×0.10×0.10 Temperature/K 291(2) 298(2) Dc/(Mg·m-3) 2.057 1.609 F(000) 830 1 220 θ range/(°) 1.678~28.454 2.35~26.32 Limiting indices -13≤h≤11,-15≤k≤14,-16≤l≤16 -20≤h≤15,-20≤k≤20,-16≤l≤17 Collected reflections 10 532 12 687 Independent reflections (Rint) 7 020(0.053 7) 3 682(0.064 9) Observed reflections [I > 2σ(I)] 3 350 2 102 Refinement method Full-matrix least squares on F2 Full-matrix least squares on F2 Goodness-of-fit on F2 1.002 0.996 Final R indices [I > 2σ(I)] R1=0.062 2, wR2=0.144 4 R1=0.056 6, wR2=0.148 7 R indices(all data) R1=0.117 2, wR2=0.155 5 R1=0, 113 1, wR2=0.179 0 Largest diff. peak and hole/(e·nm-3) 991 and -1 182 1 165 and -502 CCDC: 1437662, 1; 1014482, 2.
1.4 Oxidative carbonylation of methanol and the analysis of the product
The oxidative carbonylation of methanol with CO and O2 was conducted in a 250 mL stainless steel autoclave lined with Teflon. First, 40 mL of methanol and 0.44 mmol of the catalyst were loaded into the autoclave. Second, the air in the autoclave was displaced three times with O2, followed by pressuriza-tion to 4.0 MPa with CO and O2 (pCO/pO2=19) at room temperature. Third, the system was heated to 120 ℃ and maintained for 4 h. After the reaction, the reactor was cooled down to room temperature. Next, the reaction mixture was analyzed on a Shimadzu GC-2014 equipped with an RTX-50 capillary column (30 m×0.32 mm×0.25 μm) and a FID. The conditions employed are as follows: column temperature, 60 ℃; injector temperature, 250 ℃; detector temperature, 300 ℃; FID detection, calibration normalization method.
2 Results and discussion
2.1 Structure description of 1
The Cu complex 1 is a Cu(Ⅰ)-Cu(Ⅱ) mixed-valent complex. The unit cell structure of 1 (Fig. 1) contains two [Cu(Ⅱ)(phen)2Br]+ cations and one tetranuclear [Cu(Ⅰ)4Br6]2- anion.
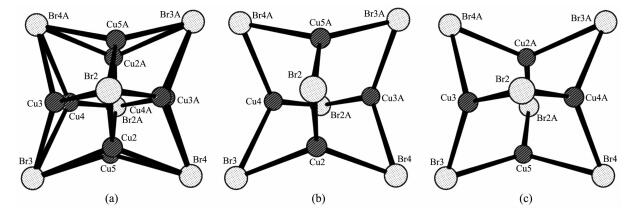
Comparison with the [Cu(Ⅱ)(phen)2Br]+ isomers reported in the literature[17], the [Cu(Ⅱ)(phen)2Br]+ cation of 1 has a square based pyramidal distorted trigonal bipyramidal (SBPDTB) stereochemistry, as the τ value, where τ=(α8-α1)/60°[18] (α8: N(4)-Cu(1)-N(1), α1: N(2)-Cu(1)-Br(1)), is 0.88. The [Cu(Ⅰ)4Br6]2- anion is a Cu(Ⅰ) tetranuclear structure composed of four monovalent copper atoms and six bromine atoms. The four copper atoms Cu(2), Cu(3A), Cu(4) and Cu(5A) in Fig. 2b exhibit approximate trigonal planar coordination with slight deviations from the planes through three bromine atoms, constituting a distorted tetrahedron with bond angles varying from 57.06(7)° to 64.05(7)° (Table 2). Because copper atoms in the [Cu(Ⅰ)4Br6]2- anion are positional disorder, the Cu(Ⅰ) tetrahedron can assume either of two equivalent orientations, as shown in Fig. 2b and 2c. Six bromine ligands lie on the six edges of the Cu(Ⅰ) tetrahedron to bridge four copper atoms; thus, the [Cu(Ⅰ)4Br6]2- anion is an agg-regate composed of an octahedron of bromide ligands containing a tetrahedron of trigonal-planar-coordinated Cu(Ⅰ) atoms.
Cu(1)-N(1) 0.199 4(6) Br(2)-Cu(3) 0.244 4(2) Br(3)-Cu(5) 0.235 0(2) Cu(1)-N(2) 0.2l3 4(6) Br(2)-Cu(4A) 0.235 9(2) Br(4)-Cu(2) 0.240 7(2) Cu(1)-N(3) 0.2l0 4(6) Br(2)-Cu(5A) 0.241 5(2) Br(4)-Cu(3A) 0.241 8(2) Cu(1)-N(4) 0.198 0(6) Br(3)-Cu(2) 0.252 9(2) Br(4)-Cu(4A) 0.238 4(2) Br(1)-Cu(1) 0.243 88(11) Br(3)-Cu(3) 0.231 1(2) Br(4)-Cu(5) 0.244 5(2) Br(2)-Cu(2) 0.233 3(2) Br(3)-Cu(4) 0.244 2(2) N(2)-Cu(1)-Br(1) 117.83(16) Br(2A)-Cu(4)-Br(3) 121.53(9) Cu(4A)…Cu(3)…Cu(5) 62.43(7) N(3)-Cu(1)-Br(1) 137.20(14) Br(2A)-Cu(4)-Br(4A) 122.95(9) Cu(5)…Cu(3)…Cu(2A) 57.07(7) N(3)-Cu(1)-N(2) 105.0(2) Br(4A)-Cu(4)-Br(3) 114.73(8) Cu(2)…Cu(4)…Cu(5A) 58.25(7) N(4)-Cu(1)-N(1) 170.9(2) Br(2A)-Cu(5)-Br(4) 115.44(8) Cu(3A)…Cu(4)…Cu(2) 64.05(7) Br(2)-Cu(2)-Br(3) 121.30(9) Br(3)-Cu(5)-Br(4) 121.12(8) Cu(3A)…Cu(4)…Cu(5A) 60.51(7) Br(2)-Cu(2)-Br(4) 123.05(9) Br(3)-Cu(5)-Br(2A) 123.09(9) Cu(2A)…Cu(5)…Cu(3) 62.70(7) Br(4)-Cu(2)-Br(3) 115.54(9) Cu(4)…Cu(2)…Cu(3A) 57.89(7) Cu(2A)…Cu(5)…Cu(4A) 58.16(7) Br(4A)-Cu(3)-Br(2) 115.32(9) Cu(4)…Cu(2)…Cu(5A) 63.59(7) Cu(3)…Cu(5)…Cu(4A) 57.06(7) Br(3)-Cu(3)-Br(2) 126.08(9) Cu(5A)…Cu(2)…Cu(3A) 60.23(7) Br(3)-Cu(3)-Br(4A) 118.47(9) Cu(4A)…Cu(3)…Cu(2A) 58.06(7) Symmetry codes: A: 1-x, -y, 1-z In the [Cu(Ⅰ)4Br6]2- anion, the copper-copper dist-ances (Cu(4)…Cu(3A) 0.268 7(3) nm, Cu(2)…Cu(4) 0.269 2(3) nm, Cu(2)…Cu(5A) 0.269 4(3) nm, Cu(3)…Cu(5) 0.278 6(3) nm) are shorter than two times the sum of the van der Waals radii of Cu atoms (0.280 nm), except for Cu(2) …Cu(3A) (0.285 2(3) nm) and Cu(4)…Cu(5A) (0.283 8(3) nm), suggesting strong Cu(Ⅰ)-Cu(Ⅰ) interac-tion[19].
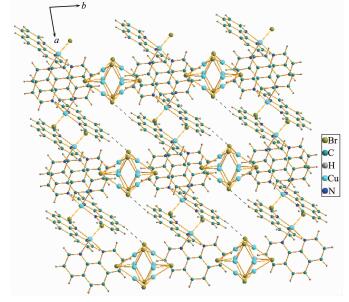
The distances of π-π interactions between the phen six-membered rings for 1 vary from 0.341 96 to 0.351 76 nm. Table 3 lists the values of the C-H…Br hydrogen bond constituting the Br atom in the [Cu(Ⅰ)4Br6]2- anion and the C atom in the offset phen ring. The structure of 1 can be described as a supra-molecular network assembled via the combination of π-π interactions and C-H…Br hydrogen bonds, in which tetranuclear Cu(Ⅰ) anions reside inside the cavities of the frameworks (Fig. 3).
D-H…A d(D-H)/nm d(H…A)/nm d(D-A)/nm ∠DHA/(°) C(24)-H(24)…Br(4B) 0.093 0.292 0.381 6(8) 162 Symmetry codes: B: -1+x, y, z 2.2 Structural description of 2
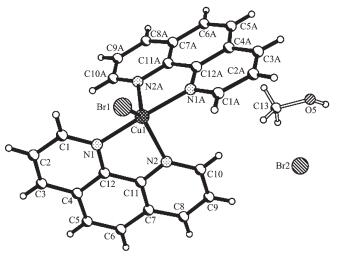
X-ray crystal-structure analysis reveals that 2 crystallizes in the C2/c space group. Table 4 lists the selected bond distances and angles of 2, and Fig. 4 and 5 summarize the asymmetry unit and packing projection of 2, respectively.
Cu(1)-N(1) 0.200 0(3) Cu(1)-N(2) 0.213 5(3) Cu(1)-Br(1) 0.241 74(11) Cu(1)-N(1A) 0.200 0(3) Cu(1)-N(2A) 0.213 5(3) N(1)-Cu(1)-N(1A) 170.27(19) N(2A)-Cu(1)-Br(1) 130.37(9) N(1)-Cu(1)-Br(1) 94.86(9) N(2)-Cu(1)-N(2A) 99.26(18) N(1)-Cu(1)-N(2) 80.49(13) N(2)-Cu(1)-Br(1) 130.37(9) N(1)-Cu(1)-N(2A) 93.18(13) Symmetry codes: A: -x, y, -z+1/2 Complex 2 consists of [Cu(Ⅱ)(phen)2Br]+, Br- and CH3OH (Fig. 4). None of the anions or methanol mole-cules are close enough ( < 0.3 nm) to be considered even weakly semi-coordinated to the Cu(Ⅱ) cation[17]. The [Cu(Ⅱ)(phen)2Br]+ cation of 2, also involves a near trigonal bipyramidal stereochemistry having a square based pyramidal distortion (SBPDTB), with τ=0.67 (α8: N(1)-Cu(1)-N(1A), α1: N(2)-Cu(1)-Br(1)). The structural data show that the Cu and Br atoms in 2 lie on a 2-fold axis of symmetry (Table 4), while those in 1 and [Cu(phen)2Br]Br·H2O[17] do not. The correspon-ding Cu-N bond distances in 2, which are in the normal range (0.199 7~0.215 0 nm)[20], are longer than those in [Cu(phen)2Br]Br·H2O[17], and comparable to those in 1. Meanwhile, the Cu-Br bond distances in the [Cu(Ⅱ)(phen)2Br]+ cations of 1 and 2, are shorter than that in [Cu(phen)2Br]Br·H2O[17].
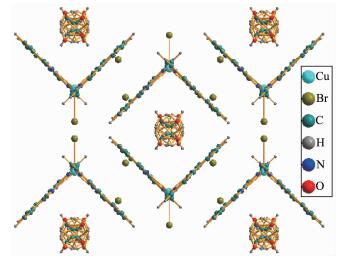
Viewing from the direction shown in Fig. 5, the most remarkable structural feature is that 2 exhibits a supramolecular framework. In 2, there are π-π stacking interactions during the six-membered rings of phen with centroid-centroid distance varying from 0.337 07 to 0.338 34 nm. The solvent methanol mole-cules in an orientational disorder state reside inside the gaps of the frameworks, indicating that methanol can evaporate easily.
2.3 TG analysis of complexes
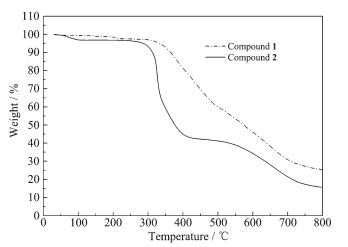
Thermogravimetric analysis of 1 shows that 1 begins to lose weight at approximately 290 ℃ (Fig. 6), indicating that 1 is stable below 290 ℃. A weight loss of 41.35% is observed in the temperature range of 290~500 ℃, which is close to the mass fraction of phen (38.38%). Taking into account that the phen unit exhibits a melting point of 99 ℃ and a boiling point of 300 ℃, the weight loss of 41.35% is attributed to the complete decomposition of phen. At temperatures greater than 500 ℃, the residue begins to lose weight.
Thermogravimetric analysis of 2 shows that a 2.84% weight loss of 2 is observed from 50 to 110 ℃, close to the half of the methanol mass fraction (5.20%). This shows that methanol in 2 is easily removed, which is consistent with the result analyzed by the single-crystal XRD. In the temperature range of 110~260 ℃, a very flat line is observed on the TG curve, with no weight loss, indicating that [Cu(phen)2Br]Br is very stable. Continuous decomposition is observed during 260~430 ℃, and a weight loss of 54.53% is close to the mass fraction of phen (58.46%), indicating that phen is completely decomposed. At temperatures greater than 430 ℃, the weight loss is ascribed to the decomposition of the residue.
2.4 Catalytic activity of complexes
Transition-metal complexes are frequently employed as important catalysts in several catalytic reaction[21], hence, the oxidative carbonylation of methanol to DMC is selected as the probe reaction, and the catalytic performances of 1 and 2 are investigated in this reaction.
As shown in Table 5, when CuBr2 was used as the catalyst, the turnover number (TON) of DMC is 5.2 with a selectivity of 67.8% for DMC; however, the selectivity for the byproduct dimethoxy methane (DMM) is up to 32.2%. On the other hand, when 2 replaced CuBr2, the TON is 5.9. The central copper atom in 2 is penta-coordinated, and the steric hindrance from phen ligands blocks the coordination of the copper with CO and methanol; hence, the activity is low. Although the increase of the TON is not clear, the selectivity to DMC is close to 100%. Notably, the TON for 1 is up to 54.7 with 97.5% selectivity of DMC, exhibiting activity and DMC selectivity higher than those reported previously for Cu(phen)Cl2[22] and (C3H7)4NBr/CuBr2[23]. Complexes 1 and 2 have the same [Cu(Ⅱ)(phen)2Br]+ cation; however, from the catalytic property of 2 in Table 5, the activity of the [Cu(Ⅱ)(phen)2Br]+ cation is very low; hence, the high activity and DMC selectivity of 1 are attributed to the [Cu(Ⅰ)4Br6]2- anion. Four copper atoms in the [Cu(Ⅰ)4Br6]2- anion are bridged by six bromine atoms (Fig. 2), and this structure might be in favor of the oxidative carbonylation of methanol[24]; thus, 1 exhibits activity very higher than 2.
3 Conclusions
In this study, 1 with a mixed-valent Cu(Ⅰ)-Cu(Ⅱ) system was synthesized from CuBr2 and phen in alcohol by autoreduction, while 2 was prepared from CuBr and phen in methanol by oxidation, both of which can assemble into supramolecular frameworks by various interactions. The [Cu(Ⅱ)(phen)2Br]+ cations of 1 and 2 have a square based pyramidal distorted trigonal bipyramidal stereochemistry, while the Cu and Br atoms in 2 lie on a crystallographic 2-fold axis of symmetry. The [Cu(Ⅰ)4Br6]2- ion in 1 is composed of an octahedron of bromide ligands containing a tetrahedron of coordinated Cu(Ⅰ) atoms, each of which exhibits approximate trigonal planar coordination and is bridged by three bromine atoms. Four copper atoms are found to be disordered, resulting in two equivalent Cu(Ⅰ) tetrahedrons observed, and there is the strong interaction between them. When both 1 and 2 were tested as catalysts for the oxidative carbonylation of methanol to DMC, 2 exhibits the TON of only 5.9, while the TON on 1 was up to 54.7.
Supporting information is available at http://www.wjhxxb.cn
-
-
[1]
胡春燕, 聂旭亮, 熊辉, 等.无机化学学报, 2014, 30(3):621-626 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?flag=1&file_no=20140324&journal_id=wjhxxbcnHU Chun-Yan, NIE Xu-Liang, XIONG Hui, et al. Chinese J. Inorg. Chem., 2014, 30(3):621-626 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?flag=1&file_no=20140324&journal_id=wjhxxbcn
-
[2]
Hirtenlehner C, Monkowius U. Inorg. Chem. Commun., 2012, 15:109-112 doi: 10.1016/j.inoche.2011.10.003
-
[3]
Gschwind F, Sereda O, Fromm K M. Inorg. Chem., 2009, 48: 10535-10547 doi: 10.1021/ic9009064
-
[4]
Wu T, Li M, Li D, et al. Cryst. Growth Des., 2008, 8:568-574 doi: 10.1021/cg070639f
-
[5]
Yang Y, Cai W, Song L, et al. Acta Crystallogr. Sect. E, 2010, 66:m1486 doi: 10.1107/S1600536810043205
-
[6]
Zhang Z Y, Deng Z P, Zhang X F, et al. CrystEngComm, 2014, 16:359-368 doi: 10.1039/C3CE41774C
-
[7]
Zhang S, Cao Y, Zhang H, et al. J. Solid State Chem., 2008, 181:3327-3336 doi: 10.1016/j.jssc.2008.09.011
-
[8]
Liu J B, Li H H, Chen Z R, et al. J. Cluster Sci., 2009, 20: 515-523 doi: 10.1007/s10876-009-0240-y
-
[9]
Sabounchei S J, Pourshahbaz M, Hashemi A, et al. J. Orga-nomet. Chem., 2014, 761:111-119 doi: 10.1016/j.jorganchem.2014.03.017
-
[10]
韩学锋, 蔡红新, 贾磊, 等.无机化学学报, 2015, 31(7):1453-1459 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?flag=1&file_no=20150727&journal_id=wjhxxbcnHAN Xue-Feng, CAI Hong-Xin, JIA Lei, et al. Chinese J. Inorg. Chem., 2015, 31(7):1453-1459 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?flag=1&file_no=20150727&journal_id=wjhxxbcn
-
[11]
Houser R P, Young V G, Tolman W B. J. Am. Chem. Soc., 1996, 118:2101-2102 doi: 10.1021/ja953776m
-
[12]
Gao X, Zhai Q G, Li S N, et al. J. Solid State Chem., 2010, 183:1150-1158 doi: 10.1016/j.jssc.2010.03.007
-
[13]
Sheldrick G M. SHELXS-97, Program for X-ray Crystal Structure Solution, University of Göttingen, Göttingen, Germany, 1997.
-
[14]
Sheldrick G M. SHELXL-97, Program for X-ray Crystal Structure Refinement, University of Göttingen, Göttingen, Germany, 1997.
-
[15]
Sheldrick G M. Acta Crystallogr. Sect. A, 2015, A71:3-8 doi: 10.1107/S2053273314026370/citedby
-
[16]
Sheldrick G M. Acta Crystallogr. Sect. C, 2015, C71:3-8 http://onlinelibrary.wiley.com/resolve/doi?DOI=10.1107%2FS2053273314026370
-
[17]
Murphy G, O'Sullivan C, Murphy B, et al. Inorg. Chem., 1998, 37:240-248 doi: 10.1021/ic970458a
-
[18]
Addison A W, Nageswara Rao T, Reedjik J, et al. J. Chem. Soc., Dalton Trans., 1984:1349-1356
-
[19]
Kim T H, Shin Y W, Kim J S, et al. Inorg. Chem. Commun., 2007, 10:717-719 doi: 10.1016/j.inoche.2007.03.009
-
[20]
毛盼东, 闫玲玲, 吴伟娜, 等.无机化学学报, 2016, 32(5):879-883 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?flag=1&file_no=20160520&journal_id=wjhxxbcnMAO Pan-Dong, YAN Ling-Ling, WU Wei -Na, et al. Chinese J. Inorg. Chem., 2016, 32(5):879-883 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?flag=1&file_no=20160520&journal_id=wjhxxbcn
-
[21]
Caballero A, Pérez P J. J. Organomet. Chem., 2015, 793:108-113 doi: 10.1016/j.jorganchem.2015.02.029
-
[22]
杜治平, 周彬, 黄丽明, 等.催化学报, 2012, 33(4):736-742 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htmDU Zhi-Ping, ZHOU Bin, HUANG Li-Ming, et al. Chin. J. Catal., 2012, 33(4):736-742 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htm
-
[23]
Raab V, Merz M, Sundermeyer J. Acta Crystallogr. Sect. E, 2001, 175:51-63 http://www.sciencedirect.com/science/article/pii/S1381116901002205
-
[24]
Liu D H, He J, Sun L B, et al. J. Taiwan Inst. Chem. Eng., 2011, 42:616-621 doi: 10.1016/j.jtice.2010.10.005
-
[1]
-
Table 1. Crystallographic data of complexes 1 and 2
Complex 1 2 Empirical formula C24H16Br4Cu3N4 C25H20Br2CuN4O Formula weight 870.67 615.81 Crystal system Triclinic Monoclinic Space group p1 C2/c a/nm 1.033 94(19) 1.428 0(4) b/nm 1.129 6(2) 1.494 4(4) c/nm 1.243 9(2) 1.243 6(4) α/(°) 78.194(4) 90.00 β/(°) 84.108(4) 106.638(5) γ/(°) 82.581(4) 90.00 V/nm3 1.405 8(5) 2.542 8(13) Z 2 4 Crystal size/mm 0.20×0.20×0.18 0.20×0.10×0.10 Temperature/K 291(2) 298(2) Dc/(Mg·m-3) 2.057 1.609 F(000) 830 1 220 θ range/(°) 1.678~28.454 2.35~26.32 Limiting indices -13≤h≤11,-15≤k≤14,-16≤l≤16 -20≤h≤15,-20≤k≤20,-16≤l≤17 Collected reflections 10 532 12 687 Independent reflections (Rint) 7 020(0.053 7) 3 682(0.064 9) Observed reflections [I > 2σ(I)] 3 350 2 102 Refinement method Full-matrix least squares on F2 Full-matrix least squares on F2 Goodness-of-fit on F2 1.002 0.996 Final R indices [I > 2σ(I)] R1=0.062 2, wR2=0.144 4 R1=0.056 6, wR2=0.148 7 R indices(all data) R1=0.117 2, wR2=0.155 5 R1=0, 113 1, wR2=0.179 0 Largest diff. peak and hole/(e·nm-3) 991 and -1 182 1 165 and -502 Table 2. Selected bond lengths (nm) and angles (°) for complex 1
Cu(1)-N(1) 0.199 4(6) Br(2)-Cu(3) 0.244 4(2) Br(3)-Cu(5) 0.235 0(2) Cu(1)-N(2) 0.2l3 4(6) Br(2)-Cu(4A) 0.235 9(2) Br(4)-Cu(2) 0.240 7(2) Cu(1)-N(3) 0.2l0 4(6) Br(2)-Cu(5A) 0.241 5(2) Br(4)-Cu(3A) 0.241 8(2) Cu(1)-N(4) 0.198 0(6) Br(3)-Cu(2) 0.252 9(2) Br(4)-Cu(4A) 0.238 4(2) Br(1)-Cu(1) 0.243 88(11) Br(3)-Cu(3) 0.231 1(2) Br(4)-Cu(5) 0.244 5(2) Br(2)-Cu(2) 0.233 3(2) Br(3)-Cu(4) 0.244 2(2) N(2)-Cu(1)-Br(1) 117.83(16) Br(2A)-Cu(4)-Br(3) 121.53(9) Cu(4A)…Cu(3)…Cu(5) 62.43(7) N(3)-Cu(1)-Br(1) 137.20(14) Br(2A)-Cu(4)-Br(4A) 122.95(9) Cu(5)…Cu(3)…Cu(2A) 57.07(7) N(3)-Cu(1)-N(2) 105.0(2) Br(4A)-Cu(4)-Br(3) 114.73(8) Cu(2)…Cu(4)…Cu(5A) 58.25(7) N(4)-Cu(1)-N(1) 170.9(2) Br(2A)-Cu(5)-Br(4) 115.44(8) Cu(3A)…Cu(4)…Cu(2) 64.05(7) Br(2)-Cu(2)-Br(3) 121.30(9) Br(3)-Cu(5)-Br(4) 121.12(8) Cu(3A)…Cu(4)…Cu(5A) 60.51(7) Br(2)-Cu(2)-Br(4) 123.05(9) Br(3)-Cu(5)-Br(2A) 123.09(9) Cu(2A)…Cu(5)…Cu(3) 62.70(7) Br(4)-Cu(2)-Br(3) 115.54(9) Cu(4)…Cu(2)…Cu(3A) 57.89(7) Cu(2A)…Cu(5)…Cu(4A) 58.16(7) Br(4A)-Cu(3)-Br(2) 115.32(9) Cu(4)…Cu(2)…Cu(5A) 63.59(7) Cu(3)…Cu(5)…Cu(4A) 57.06(7) Br(3)-Cu(3)-Br(2) 126.08(9) Cu(5A)…Cu(2)…Cu(3A) 60.23(7) Br(3)-Cu(3)-Br(4A) 118.47(9) Cu(4A)…Cu(3)…Cu(2A) 58.06(7) Symmetry codes: A: 1-x, -y, 1-z Table 3. Hydrogen bond parameters for complex 1
D-H…A d(D-H)/nm d(H…A)/nm d(D-A)/nm ∠DHA/(°) C(24)-H(24)…Br(4B) 0.093 0.292 0.381 6(8) 162 Symmetry codes: B: -1+x, y, z Table 4. Selected bond lengths (nm) and angles (°) for complex 2
Cu(1)-N(1) 0.200 0(3) Cu(1)-N(2) 0.213 5(3) Cu(1)-Br(1) 0.241 74(11) Cu(1)-N(1A) 0.200 0(3) Cu(1)-N(2A) 0.213 5(3) N(1)-Cu(1)-N(1A) 170.27(19) N(2A)-Cu(1)-Br(1) 130.37(9) N(1)-Cu(1)-Br(1) 94.86(9) N(2)-Cu(1)-N(2A) 99.26(18) N(1)-Cu(1)-N(2) 80.49(13) N(2)-Cu(1)-Br(1) 130.37(9) N(1)-Cu(1)-N(2A) 93.18(13) Symmetry codes: A: -x, y, -z+1/2 -

 扫一扫看文章
扫一扫看文章
计量
- PDF下载量: 1
- 文章访问数: 751
- HTML全文浏览量: 77



 下载:
下载:





 下载:
下载:

